Figures & data
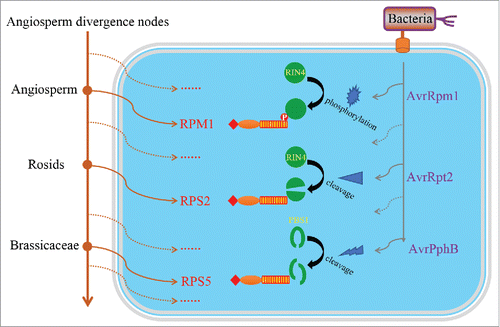
Figure 1. The evolutionary history of 3 functional R genes against Pseudomonas syringae. To evade the activation of plant immune system, bacteria have evolved various effector proteins to modify or disrupt certain important host immune-related proteins. As an early-originating R gene, RPM1 specifically detects the phosphorylation of RIN4, which is caused by the P. syringae effector protein AvrRpm1. P. syringae then evolved another effector (AvrRpt2) to evade the activation of RPM1 by cleaving the RIN4 protein. In response, plants further evolved RPS2 to monitor the cleavage of RIN4 and activate the immune system. On the other hand, P. syringae evolved AvrPphB to target another host protein PBS1 for cleavage, which is subsequently recognized by a later evolved R gene RPS5. Solid circles indicate divergence nodes in angiosperm clade. Solid arrows indicate the origin of R genes (in brown) or bacterial effector proteins (in gray), whereas dashed arrows indicate unknown R genes or effector proteins that developed during its evolutionary history.