Figures & data
Figure 1. Phenotype differences between maize seedlings colonized and uncolonized by P. indica. (A) Typical root architecture of maize 15 days after inoculation (left) or uninoculation (B, right) with P. indica, P. indica chlamydospores on maize tap root inspected by Trypan blue staining (white arrow) at 15 DAI under microscope (Leica DM5000 B, Germany), with scale bar=20 μm. (C, D) Typical shoot and root phenotype of maize 15 days after inoculation (right) or uninoculation (left) with P. indica, with scale bar=5 cm. (E-H): Length of tap root (E), fresh weight (FW) of roots (F), dry weight (DW) of roots (G), and leaf area (H) of maize seedlings inoculated or uninoculated with P. indica under drought stress, while uninoculation and not-drought stress-exposed seedlings were used as control. All experiments were performed with at least 3 replications. Statistically significant differences are labeled with letters (p< 0.05, two-wayfactor ANOVAone way ANOVA).
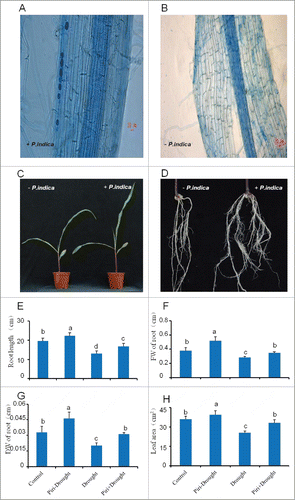
Figure 2. Experiment design and expression correlation between maize genes in roots (A) Strategies to decipher P. indica-induced drought tolerance related genes. (B) Clustering of different samples by spearman correlation analysis based on FPKM (Fragment Per Kilo bases per Million reads) of each transcript. The color scale indicates the degree of correlation coefficient (blue for low correlation and pink for high correlation). Samples clustered together were indicated by rectangle box (red, blue, and green, respectively). The map was generated using R packages.
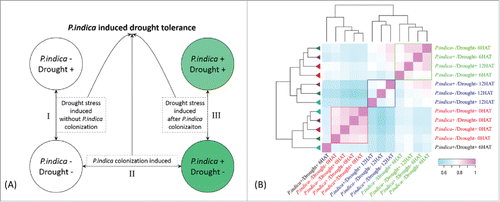
Table 1. Statistics of transcripts identified from different treatments.
Table 2. Comparison of DEGs among different treatments.
Table 3. Statistics of DEGs and gene ontology enrichment from different treatments.
Figure 3. Classification of drought induced differentially expressed genes (DEGs) 1337 DEGs were obtained by comparing transcripts identified from P. indica-/Drought+ and P. indica-/Drought-. (I in ) (A) Average linkage clustering of drought-regulated DEGs by HCL (hierarchical clustering), where log2 (fold change) less than 1.6 were not shown. (B) Expression pattern of drought regulated DEGs by K-means clustering, where each line represents a single gene and the pink line indicates the general trend within a cluster. Significant GO terms (Chi-square test with p<0.01 and FDR<0.05) were presented for each DEGs cluster, with P and F indicating biological processes and molecular functions, respectively.
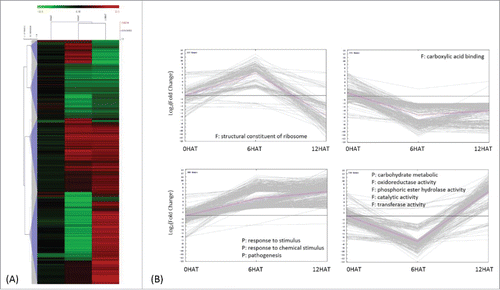
Figure 4. Classification of genes regulated by P. indica colonization. 464 DEGs were identified by comparing transcripts from P. indica+/Drought- and P. indica-/Drought-. (II in ) (A), (B) same as . (C) Deposition of catalytic activity (B) related genes by GO enrichment analysis, and the graph was automatically generated by agriGO.
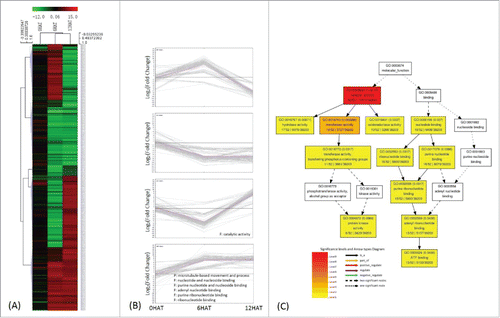
Figure 5. Classification of drought-regulated genes in P. indica colonized maize seedlings 2037 DEGs regulated by drought were identified by comparing transcripts from P. indica+/Drought+ and P. indica+/Drought- (III in ). (A) and (B) same as . (C) Detailed GO classification of cluster 3 in (B). Top ten terms in biological processes and molecular functions were shown, respecctively.
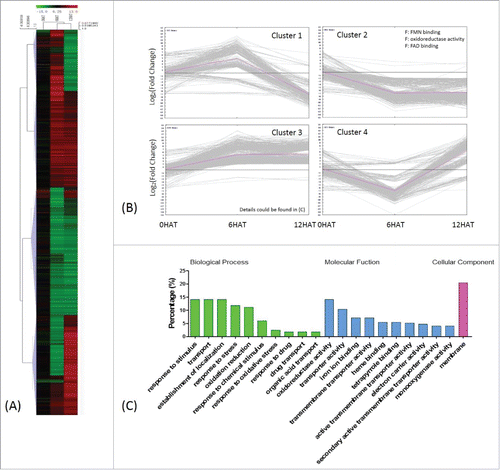
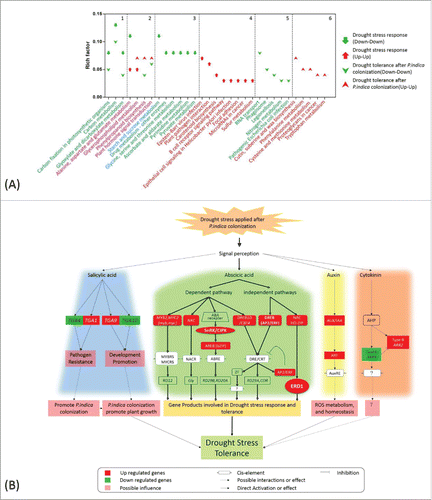
Figure 6. KEGG pathway enrichment and schematic diagram of hormone-related genes involved in drought stress tolerance. (A) DEGs detected under different conditions were analyzed by KEGG mapper, and enrichment was performed by hypergeometric probability distribution. Pathways with p<0.01 and FDR<0.05 are shown. 1 and 2: pathways commonly involved in drought stress response and drought tolerance after P. indica colonization; 3-6: pathways specific to DEG sets as denoted in the figure. (B) Schematic diagram of hormone–related genes involved in drought stress tolerance based on KEGG mapping of hormone-related DEGs, relative knowledge documented in reference (detailed annotation and expression pattern of each gene is found in the supplemental tables).