Figures & data
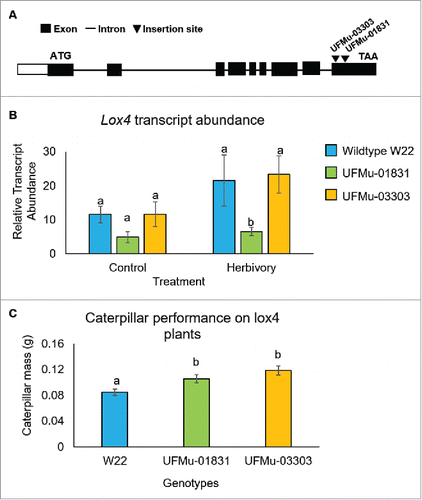
Figure 1. 9-Lipoxygenase and 13-lipoxygenase oxygenate α-linolenic acid at different positions to produce (10E,12Z)-9-Hydroperoxy-10,12,15-octadecatrienoic acid (9-HPOT and (10E,12Z)-13-hydroperoxy-10,12,15-octadecatrienoic acid (13-HPOT), respectively. 9-HPOT and 13-HPOT serve as precursors for differing sets of bioactive plant metabolites. 10-OPDA = 10-oxophytodienoic acid;12-OPDA = 12-oxophytodienoic acid; DES = divinyl ether synthase; AOS = allene oxide synthase; HPL = hydroperoxide lysase. Modified from Schiller et al. (2015).
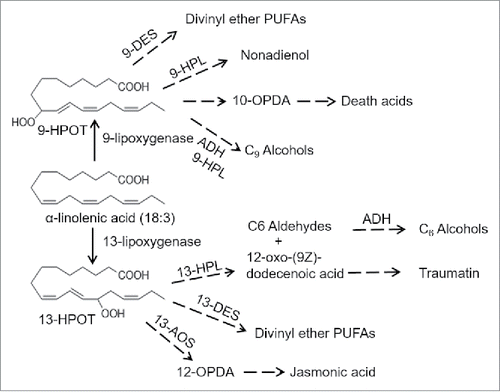
Figure 2. Dendrogram of predicted protein sequences of 27 lipoxygenases from the maize inbred line B73 v4 assembly (Zm00001d numbers) and the inbred line W22 v2 assembly (Zm00004b numbers). The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model. Evolutionary analyses were conducted in MEGA7. The tree with the highest log likelihood is shown. Bootstrap values are based on 1000 replications. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions with less than 80% site coverage were eliminated. There were a total of 835 positions in the final dataset. Proteins that are truncated and likely non-functional in W22 are marked with asterisks.
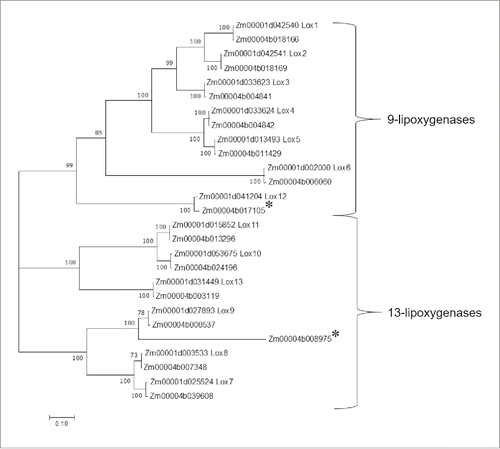
Figure 3. Caterpillar-induced expression of 9-lipoxygenases (blue bars) and 13-lipoxygenases (orange bars) in maize inbred line B73. Mean +/- s.e. of 4 independent samples. *P value < 0.05, Dunnett's test relative to 0-hour controls. Data are adapted from Tzin et al. 2017.
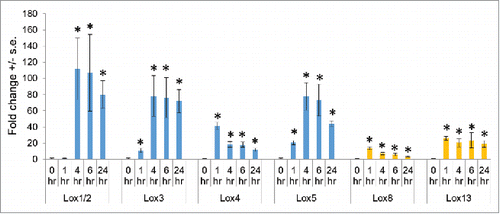
Figure 4. Analysis of a W22 Lox4 Mu insertion lines (A) Location of the UFMu-01831and UFMu-03303 insertions in the Lox4 gene and positions of primers that were used for quantitative RT-PCR gene expression analysis. (B) Expression level of the Lox4 gene, with and without six hours of S. exigua feeding. Mean +/- s.e. of 5 independent samples. (C) Mass of S. exigua caterpillars after five days of feeding on W22, UFMu-01831, and UFMu-03303seedlings. Mean +/- s.e. of 57 samples. *P value < 0.05, ANOVA followed by Tukey's HSD test.