Figures & data
Figure 1. KAI2 signaling regulates light-dependent germination.
(a) Germination assay of wild type (WT) and ply2 mutant under PhyB-dependent condition as depicted in the diagrams. After sterilization, wild type (WT, Col-0) and the mutant seeds were placed in aqueous media. After irradiation with far-red light (1.33 μW cm−2) for 15 min, and then without (PhyBoff) or with subsequent red light (5.72 μW cm−2) for 10 min (PhyBon), the plates were incubated in darkness. After 4 days, the germination rate was determined based on seedling establishment. Average of germination rate was derived from experimental triplicates with at least 100–200 seeds per plate, respectively. Error bars represent SDs. (b) Gene expression analysis. Total RNA was extracted from 12 h imbibed seeds that were treated as in (A) and subjected to quantitative real-time RT-PCR analysis. PP2A was used as an internal control. Values in the graph represent means ± SDs of the relative expression level of the denoted gene, normalized to that of PP2A (n = 3). Different letters indicate significant differences at P< 0.01, analyzed by one-way ANOVA with a post hoc Tukey HSD test. Part of data for the PhyBon experimental set were reproduced from Lee et al. (2018).
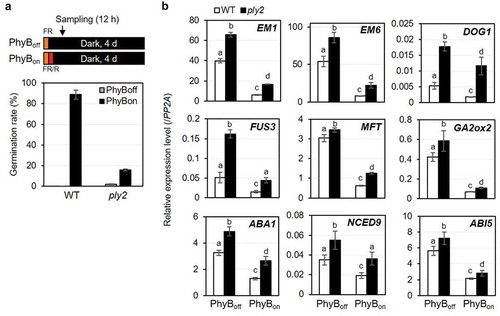
Figure 2. Double mutant analysis of ply2 and cop1-6 mutations.
(a) Representative seedling phenotypes. After irradiation with white light for 12 h to stimulate germination, the seedlings were grown on half strength MS media with or without 10 μM of KAR2 (+KAR) for 5 days in darkness. The images show representative 5 d-old dark-grown seedlings. Scale bar, 5 mm. WT, wild type. (b) Quantitative measurement of hypocotyl length. Values represent means ± SDs of hypocotyl length from at least 15 seedlings grown as in (A). WT, wild type. Asterisks indicate significant differences at p < 0.01 (Student t-test).
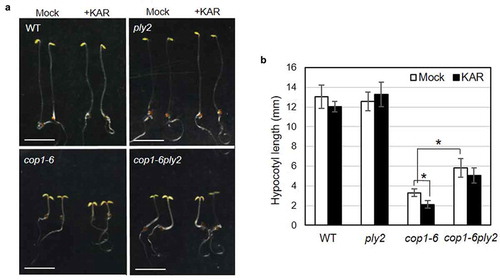
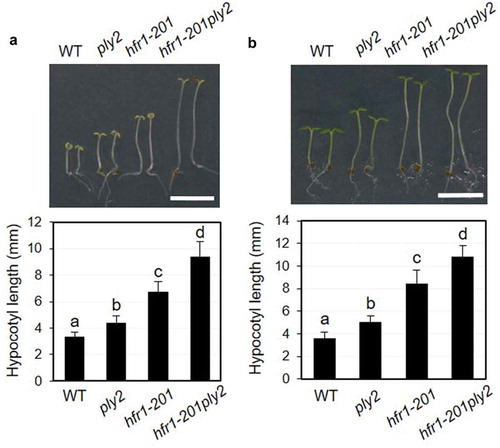
Figure 3. Double mutant analysis of ply2 and hfr1-201 mutations.
(a) Hypocotyl growth phenotypes under continuous far red light. The seedlings were grown on half strength MS media under far red light (1.33 μW cm−2) for 6 days. Upper, Representative seedling phenotypes. Scale bar, 5 mm. Lower, Values represent means ± SDs of hypocotyl length (n = 15–16). (b) Hypocotyl growth phenotypes under continuous blue light. The seedlings were grown on half strength MS media under blue light (17.4 μW cm−2) for 6 days. Upper, Representative seedling phenotypes. Scale bar, 5 mm. Lower, Values represent means ± SDs of hypocotyl length (n = 12–14). WT, wild type. Different letters indicate significant differences at p< 0.01, analyzed by one-way ANOVA with a post hoc Tukey HSD test.