Figures & data
Figure 1. Expression of VAMP727, SYP22, and PR1 to M. incognita. The red lines (RNK) were observed in primary root tip (a) and lateral root apex (b). Bar = 100 µm. (c) Schematic showing M. incognita inoculation and time points for sampling expression of M. incognita-responsive genes. Col-0 plants were grown for 2 weeks, followed by inoculation of M. incognita. The whole roots were harvested after 0, 12, 24, 36, 48 and 72 h of the inoclution. (d) Expression levels of VAMP727, SYP22, and PR1 genes were examined using qRT–PCR. mRNA levels in the samples were normalized against those of Actin mRNA. Data are means ± s.e. (n = 3); different letters indicate significant differences between results (P < .05).
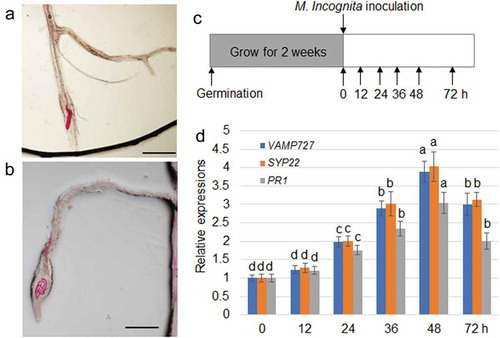
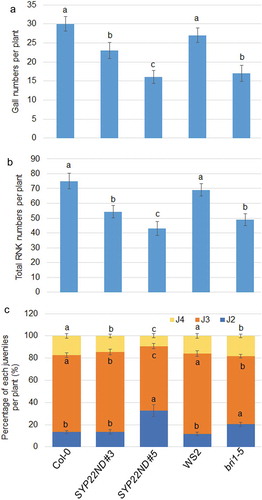
Figure 2. The numbers of gall, total RKN and each juvenile percentages of M. incognita in SYP22ND and BR insensitive mutants. (a) Number of galls in WT (Col-0 for SYP22ND mutants, WS2 for bir1-5), SYP22ND #3, #5, and bri1-5 was calculated in each plant. The error bars mean ± SE (n = 20). (b) Total number of RKN in WT, SYP22ND #3, #5, and bri1-5 was calculated in each plant. The error bars mean ± SE (n = 20). (c) The percentage of each juveniles (J2, J3, and J4) was calculated in WT, SYP22ND #3, #5, and bri1-5 was calculated in each plant. The error bars mean ± SE (n = 20). Significant differences between groups, were analyzed via one-way analysis of variance (ANOVA), followed by Bonferroni’s multiple comparison tests. Differences among samples were considered significant at P < .05.