Figures & data
Figure 1. The biosynthesis of 2-phenylethanol and 2-phenylethyl-β-D-glucopyranoside in poplar. AADC, aromatic amino acid decarboxylase; AAS, aromatic aldehyde synthase; MAO, monoamine oxidase; PAR, phenylacetaldehyde reductase; CYP79, cytochrome P450 family 79 enzyme; AAAT, aromatic amino acid transaminase; TOX, transoximase; PPDC, phenylpyruvic acid decarboxylase; UGT, UDP-glucosyl transferase; β-Glu, β-glucosidase. Dashed lines indicate enzymes/reactions not yet characterized in planta. Solid lines indicate characterized poplar enzymes, and dotted lines indicate enzymes characterized in other plants.
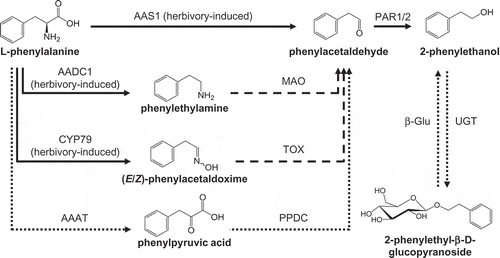
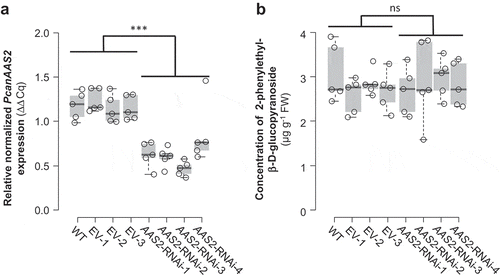
Figure 2. Transcript accumulation of PcanAAS2 (a) and accumulation of 2-phenylethyl-β-D-glucopyranoside (b) in undamaged leaves of Populus x canescens wild type plants (WT), empty vector control plants (EV), and PcanAAS2 RNAi lines (AAS2-RNAi). (a) Gene expression was analyzed using real time-quantitative PCR and the relative normalized expression compared to the reference gene ubiquitin is shown. (b) 2-Phenylethyl-β-D-glucopyranoside was extracted with methanol from ground leaf material and analyzed via liquid chromatography-tandem mass spectrometry. Biological replicates (nb) and technical replicates (nt) of EV lines and RNAi lines were used to test for statistical differences. WT, nb = 5; EV, nb = 3, nt = 5; AAS2-RNAi, nb = 4, nt = 5 (AAS2-RNAi-2, nb = 4, nt = 4). Asterisks indicate statistical significance as assessed by Student’s t tests. PcanAAS2 expression (P < 0.001, t = 8.934); 2-phenylethyl-β-D-glucopyranoside accumulation (P = 0.792, t = −0.266). Medians ± quartiles and outliers are shown. Each data point is represented by a circle. ns, not significant.