Figures & data
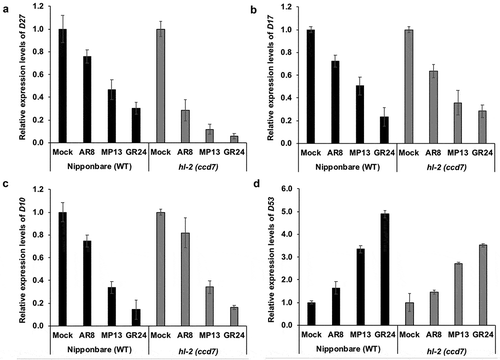
Figure 1. Biosynthesis, perception and signaling pathways of strigolactones (Adapted and modified fromCitation6). SL biosynthesis involves sequential actions of an isomerase DWARF27 (D27) and two CAROTENOID CLEAVAGE7Citation7 (CCD7) and CCD8, which take place within plastids, and Cytochrome P450 (711 clade) and other enzymes in the cytosol (red enzymes on solid arrows). D27 catalyzes the reversible isomerization of all-trans- into 9-cis-β-carotene that is converted by CCD7 and CCD8 into carlactone, a key intermediate in SL biosynthesis. Cytochrome P450 enzymes (MAX1) in the cytosol further convert carlactone into non-canonical and canonical SLs. The F-box D3 (MAX2) and D14 proteins are required for SL perception and signal transduction (dashed arrow), which triggers SL response. The transcriptional repressors SMXLs/D53 further involves in the polyubiquitination of D14 and SMXLs/D53 and in the 26S proteasomal degradation (dashed arrow).
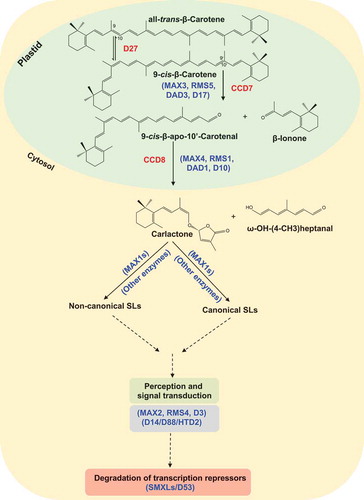
Figure 2. (a) Structure of AR8, a previously described SL analogCitation22 containing a methyl group at the C-3ʹ atom of the D-ring. (b) Structure of the SL analog MP13 that differs from AR8 by the absence of C-3ʹ methylationCitation23 (c) Structure of the common SL analog rac-GR24 consisting of two enantiomers (as shown).
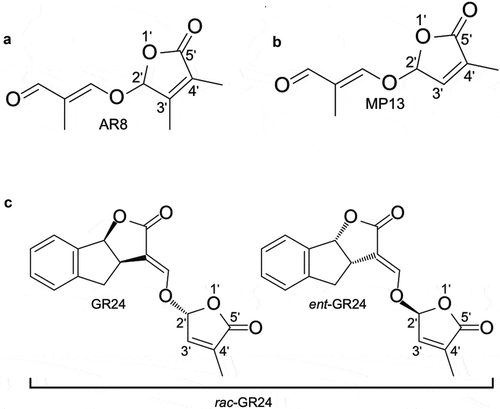
Figure 3. Effect of the treatment with AR8, MP13 and GR24 on transcript levels of the SL biosynthesis genes D27, D17, D10 (a–c) and the transcriptional repressor D53 (d). Quantitative real-time PCR measurement was performed with total RNA isolated from roots of Nipponbare (WT) and hl-2 (ccd7 mutant) rice seedlings grown under phosphorus-deficient conditions. Ubiquitin was used as the reference gene, and the transcript levels in the untreated control (mock) were normalized to 1. Bars represent means ± SD (n= 3).