Figures & data
Figure 1. Confirmation of transgenic tobacco plants.
(a) Genomic PCR analysis and histochemical GUS staining of transgenic lines. (b) Quantitative real-time PCR. Data are means ± SDs (n = 3), P < .05. Significant differences are indicated by different letters above the bars.
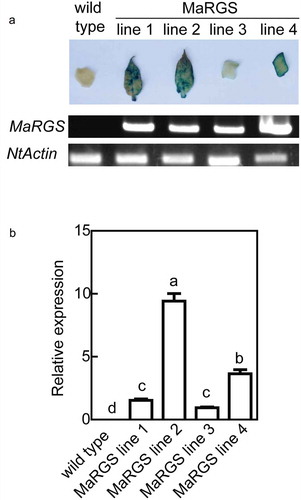
Figure 2. Salt stress tolerance analysis of MaRGS transgenic tobacco plants.
(a) The root growth of MaRGS transgenic tobacco and WT plants under normal and stress conditions. (b) The growth of MaRGS transgenic tobacco and WT plants under normal and stress conditions. (c) Statistical analysis of root lengths of MaRGS transgenic tobacco and WT plants. Data are means ± SDs (n = 15), *P < .05. (d–f) The H2O2 content (d), MDA content (e) and proline content (f) in MaRGS transgenic tobacco and WT plants under normal and stress conditions. Data are means ± SDs (n = 3), *P < .05.
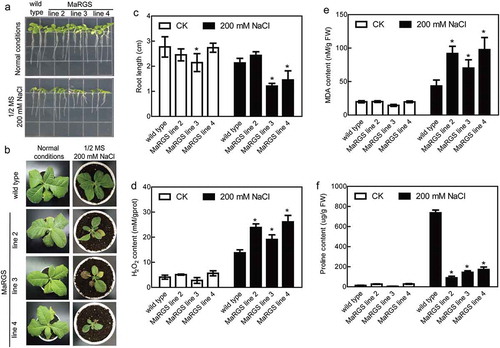
Figure 3. Salt stress analysis of MaRGS in mulberry by transient expression.
(a) Analysis of the expression of MaRGS in the control, MaRGS-overexpressing and RNAi-silenced MaRGS plants. (b,c) The H2O2 content (b) and proline content (c) in the control, MaRGS-overexpressing and RNAi-silenced MaRGS plants. Data are means ± SDs (n = 3), P < .05. Significant differences are indicated by different letters above the bars
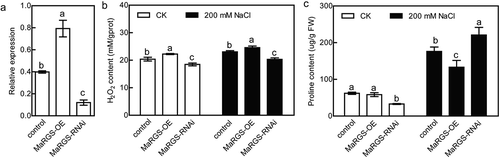
Figure 4. The soluble sugar content and the expression levels of ATG and WNK genes in mulberry and tobacco.
(a,b) The soluble sugar contents in mulberry (a) and tobacco (b) after NaCl treatment. (c,d) The expression levels of WNK and ATG genes of mulberry (c) and tobacco (d) in response to NaCl and D-glucose treatments. (e) The fold change of MaWNK1, MaWNK8, MaATG8 and MaATG7 expression in the mulberry seedlings transiently transformed with pFGC:MaRGS-OE and pFGC:MaRGS-RNAi after D-glucose treatment. (f) The fold change of NtATG8f, NtATG6, NtATG8a, NtWNK8 and NtWNK9 expression in the transgenic tobacco plants overexpressing MaRGS after D-glucose treatment. Data are means ± SDs (n = 3), *P < .05.
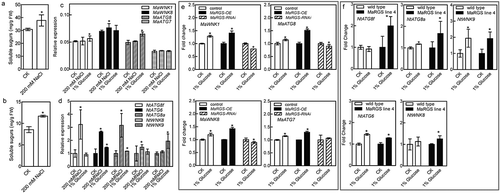
Figure 5. A possible model for RGS-regulated G-protein signaling in response to salt stress.
(a) Under normal conditions, the inactivated and activated states of G-proteins are balanced, and Gα and Gβγ subunits perform negative and positive roles, respectively, in plant response to salt stress. (b) When plants are exposed to salt stress, the accumulation of D-glucose and ATG proteins are induced, and WNK proteins are also recruited. This leads to the endocytosis of RGS and the dissociation of heterotrimeric G-proteins. Gβγ dimers eventually promote plant tolerance to salt stress.