Figures & data
Figure 1. PvRXLR159 is induced in the early stage of infection.
A, Representative photos of grapevine leaf discs inoculated with P. viticola. B, The growth of P. viticola in grapevine. The growth of P. viticola was monitored by qPCR and evaluated as the relative quantity of PvActin (P. viticola Actin) to VvActin (V. vinifera Actin). C, Transcription of PvRXLR159 in P. viticola during infection. Transcription levels of PvRXLR159 were monitored by qPCR and evaluated as the relative quantity of PvRXLR159 to PvActin. Error bars represent the standard errors from three biological replicates. hpi, hours post-inoculation.
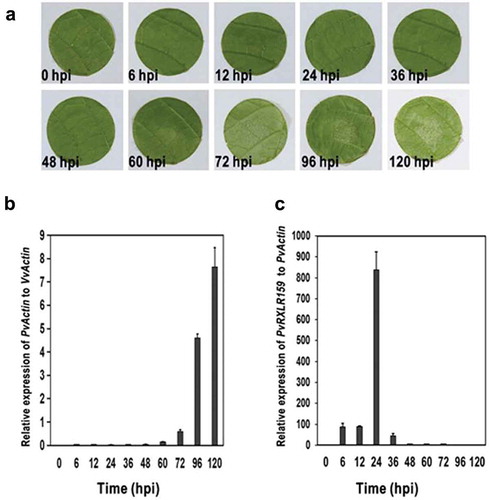
Figure 2. PvRXLR159 is an effector containing functional signal peptide.
Functional validation of the signal peptide of PvRXLR159 was carried out by using signal sequence trap assay. The signal peptide sequences of PvRXLR159 and Avr1b as a positive control, and the sequence of the first 25 amino acids of Mg87 as a negative control were fused in frame to the invertase gene in the pSUC2 vector, which were transformed into yeast YTK12 strain. CMD-W medium were used to screen positive transformants. YPRAA medium and TTC reducing assay were used to test invertase secretion. Only clones having a functional signal peptide can grow on YPRAA medium and reduce TTC to red formazan.
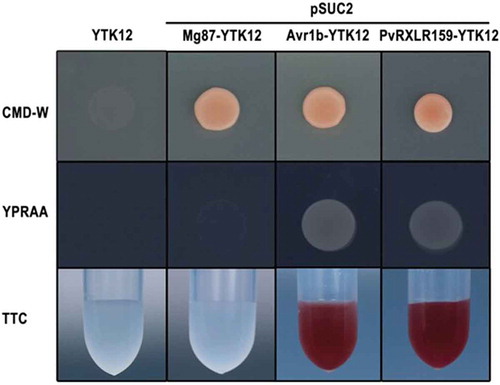
Figure 3. C terminus of PvRXLR159 is essential for its function.
Deletion mutants of PvRXLR159 were expressed in N. benthamiana leaves for 24 h, followed by expression of the elicitors INF1 and BAX to induce cell death. Left panel, a schematic diagram of the different deletion constructs. Right panel, typical symptoms of N. benthamiana taken at 5 days after expression of INF1 and BAX. The experiments were repeated at least three times with similar results.
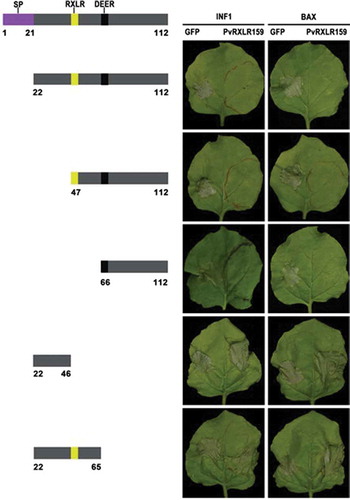
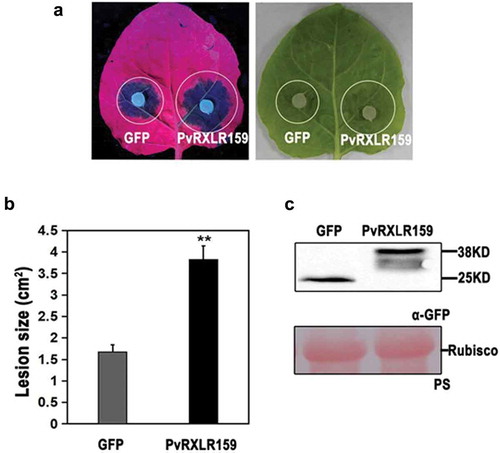
Figure 4. PvRXLR159 suppresses N. benthamiana resistance to P. capsici.
PvRXLR159-GFP and GFP were transiently expressed in N. benthamiana. Leaves were detached at 48 h, followed by inoculation with P. capsici. Images of typical symptoms were taken at 3 days after inoculated with P. capsici as shown in A. Quantification of lesion size in A was shown in B. Error bars represent the standard errors from three biological replicates. Asterisks represent significant differences from GFP (**P < .01, Student’s t-test). Expression of PvRXLR159-GFP was checked by western blot as shown in C. PS, Ponceau S staining.