Figures & data
Figure 1. Expression of C/VIF1 in response to ABA treatment. (a) Expression of C/VIF1 was induced by ABA. Seven-day-old Col seedlings were treated with 50 µM ABA or mock-treated with solvent methanol for 4 h. Total RNA was isolated, cDNA was synthesized and used as a template for RT-PCR. The expression of ACT2 was used as a control. (b) Fold changes of C/VIF1 in response to ABA. Expression level of C/VIF1 was examined by qRT-PCR. The expression of ACT2 was used as an inner control. The fold change was calculated by comparing the expression levels of C/VIF1 in ABA treated and mock-treated seedlings. Data represent the mean ± SD of three replicates.
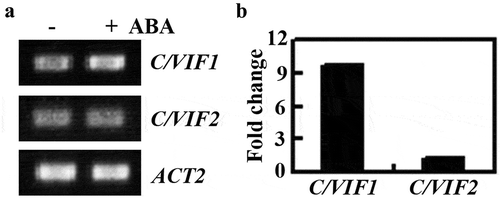
Figure 2. Generation of pHEE-FT CRISPR/Cas9 constructs for C/VIF1 gene editing. (a) Diagram of the pHEE-FT vector with a GmFT2a expression cassette. (b) The sgRNA expression cassettes in the pHEE-FT-C/VIF1 and pHEE-FT-C/VIF1-C/VIF2 constructs. The target sequences of C/VIF1 and C/VIF2 were introduced into the sgRNA expression cassettes by PCR amplification, and the sgRNA expression cassettes were cloned into the pHEE-FT vector by using Golden gate cloning. (c) Target sequences in C/VIF1 and C/VIF2. Numbers indicate the positions of the nucleotides in the coding sequences of C/VIF1 or C/VIF2. PAM sites were indicated in the brackets.
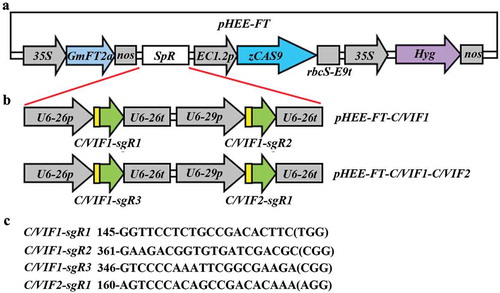
Figure 3. Isolation of transgene-free c/vif1 mutants. (a) Editing status of C/VIF1 in the c/vif1 mutants. The c/vif1-c1 and c/vif1-c2 mutants were obtained by transforming Col wild-type plants with pHEE-FT-C/VIF1-C/VIF2 and pHEE-FT-C/VIF1 constructs, respectively. Editing status in T1 generation was examined, and transgene-free homozygous mutants were isolated in T2 generation. Star indicates the single T nucleotide insertion in the c/vif1-c1 mutant. Arrowhead indicates the 216 bp fragment deletion in the c/vif1-c2 mutant. Underlines indicate the PAM sites. (b) Alignment of C/VIF1 amino acid sequences in the Col wild type and the c/vif1 mutants. ORF of C/VIF1 in the c/vif1 mutants was identified by using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) and predicted amino acid sequences were used for alignment with the amino acid sequence of C/VIF1. Underline indicates the PKF motif. (c) NBT staining of the seeds. Seeds of the Col wild type and the c/vif1 mutants were fixed with fixation buffer for 1 h at 4°C, rinsed overnight with water to remove soluble sugar, and stained with staining buffer in a water bath at 30°C. Stained seeds were rinsed with water and photographed under a dissection microscopy equipped with a digital camera.
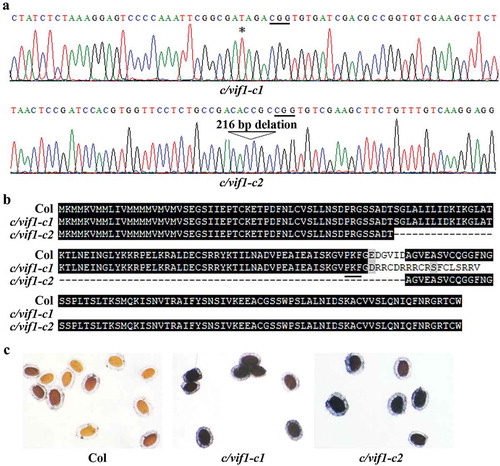
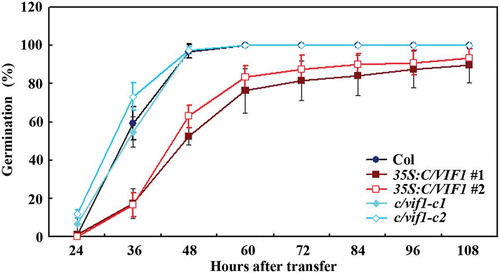
Figure 4. Effects of ABA on seed germination and cotyledon greening of the Col wild type, the 35S:C/VIF1 transgenic plants and the c/vif1 mutants. (a) Effects of ABA on seed germination of the Col wild type, the 35S:C/VIF1 transgenic plants and the c/vif1 mutants. Sterilized seeds of the Col wild type, the 35S:C/VIF1 transgenic plants and the c/vif1 mutants were germinated on 1/2 MS plates in the presence or absence of 0.75 µM ABA. The plates were transferred to a growth room after kept in darkness for 2 d at 4°C. Seeds germinated were counted every 12 h and the percentage of germination was then calculated. All the seeds on the control plates were generated 24 h after the transfer. Data represent the mean ± SD of three replicates. (b) Effects of ABA on cotyledon greening of the Col wild type, the 35S:C/VIF1 transgenic plants and the c/vif1 mutants. Sterilized seeds of the Col wild type, the 35S:C/VIF1 transgenic plants and the c/vif1 mutants were germinated on 1/2 MS plates in the presence or absence of 0.75 µM ABA. The plates were transferred to a growth room after kept in darkness for 2 d at 4°C, and pictures were taken 8 d after the transfer. (c) Percentage of green seedlings of the Col wild type, the 35S:C/VIF1 transgenic plants and the c/vif1 mutants in response to ABA treatment. Seedlings with green cotyledons were counted 8 d after the transfer and used to calculate the percentage of green cotyledons. Data represent the mean ± SD of three replicates.
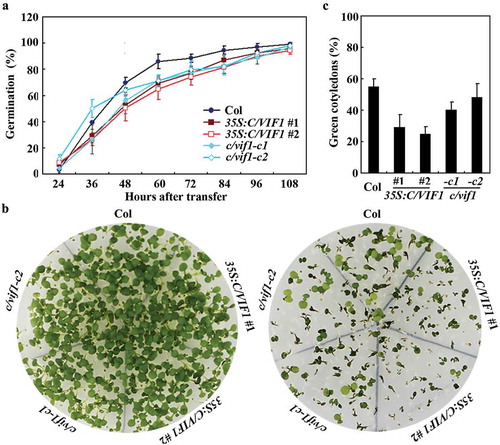
Figure 5. Effects of NaCl on seed germination of the Col wild type, the 35S:C/VIF1 transgenic plants and the c/vif1 mutants. Sterilized seeds of the Col wild type, the 35S:C/VIF1 transgenic plants and the c/vif1 mutants were germinated on 1/2 MS plates in the presence or absence of 200 mM NaCl. The plates were transferred to a growth room after kept in darkness for 2 d at 4°C. Seeds germinated were counted every 12 h and the percentage of germination was then calculated. All the seeds on the control plates generated 24 h after the transfer. Data represent the mean ± SD of three replicates.