Figures & data
Figure 1. The leaf protein amount of AOX (a) and COXII (b) in Nicotiana tabacum germinated and grown at ACO2 (400 ppm) or ECO2 (1000 ppm). In each growth condition, leaf 5 was sampled at six different days after potting. These sampling times were chosen so that leaf 5 was of comparable size in the two growth conditions at each sampling time. For example, leaf 5 of ACO2-grown plants at Day 11 was comparable in size to leaf 5 of ECO2-grown plants at Day 9. All protein amounts were normalized to the protein amount in the ACO2-grown plant at Day 11, which was set to 1. Data are the mean ± SEM of three independent experiments (n = 3). For each independent experiment and treatment, protein was isolated from leaf tissue pooled from three replicate plants. Protein was analyzed by Western blot using antibodies AS04054 (AOX1/2) and AS04053A (COXII) (Agrisera, Vännäs, Sweden). Data were analyzed by one-way ANOVA, followed by a Tukey multiple comparison test, comparing all pairs within a growth condition (ACO2 or ECO2). Data bars not sharing a common letter are significantly different from one another (P < .05).
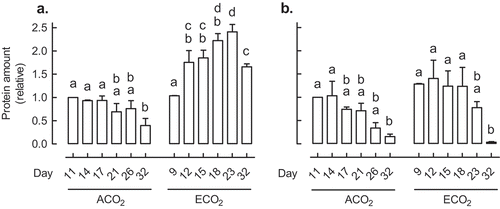
Figure 2. A phylogenetic tree of dicot GPT proteins, constructed using the maximum-likelihood method within MEGA-X software.Citation48 Protein sequences were aligned using the MUSCLE algorithm and the LG+F substitution model was used to determine branch lengths. The numbers shown on the branches are bootstrap values (performed 1000 repeats). Rooting of the tree used a monocot (Oryza sativa) GPT1 protein sequence.

Table 1. A summary of factors altering leaf GPT2 (Arabidopsis thaliana, Glycine max) or GPT3 (Nicotiana tabacum) gene expression. See text for further details.
Figure 3. The Nicotiana tabacum leaf transcript amount for genes encoding AOX (gray bars), GPT1 (black bars) and GPT3 (open bars). Plants were germinated at ACO2 (400 ppm) or ECO2 (1000 ppm) for two weeks and then potted and grown for an additional 15 days under their original growth condition, prior to either transcript analysis or transfer to an alternate growth condition. The left-most set of data (ACO2 to ECO2) shows the transcript amount 24 h after transfer of ACO2-grown plants to ECO2, compared to the amount measured in control ACO2-grown plants. The center set of data (ECO2) shows the transcript amount in ECO2-grown plants, compared to the amount measured in control ACO2-grown plants. The right-most set of data (ECO2 to ACO2) shows the transcript amount 24 h after transfer of ECO2-grown plants to ACO2, compared to the amount measured in control ECO2-grown plants. All transcript amounts were normalized to that of a reference gene (EF1) and are relative to the control amount (log2 fold-change). Data are the mean ± SEM of three independent experiments (n = 3). For each independent experiment and treatment, RNA was isolated from leaf tissue (leaf 5) pooled from three replicate plants. RNA was analyzed by qPCR, using the primers listed in Table S1.
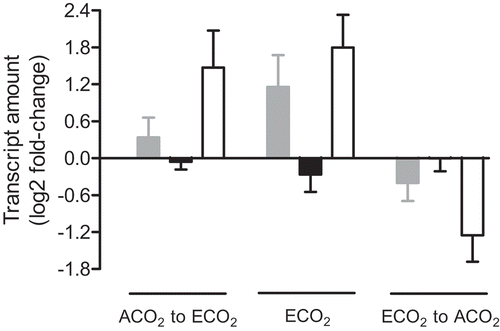
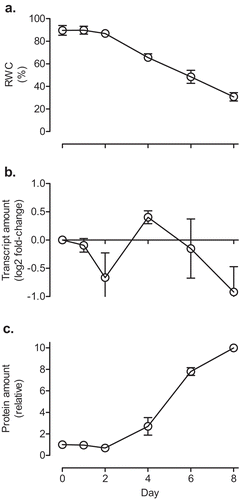
Figure 4. The relative water content (a), GPT3 transcript amount (b) and AOX protein amount (c) of Nicotiana tabacum (leaf 5) in response to water deficit. Plants were grown at ACO2 and were watered daily for 21 days. Plants were then given a final watering (Day 0 on the graph), after which time water was withheld. The GPT3 transcript amounts were normalized to that of a reference gene (GPT1) and are relative to the amount in the well-watered (Day 0) plants (log2 fold-change). GPT1 was used as the reference gene (rather than EF1, as used in ) since GPT1 amounts remained stable in response to water deficit, while EF1 amounts did not. The AOX protein amounts were normalized to the protein amount in the well-watered (Day 0) plants, which was set to 1. Data are the mean ± SEM of three independent experiments (n = 3). For each independent experiment and treatment, leaf tissue pooled from three replicate plants was used for RNA and protein isolation, as well as determination of relative water content. Protein and RNA were analyzed by Western blot and qPCR, respectively, as described in and .