Figures & data
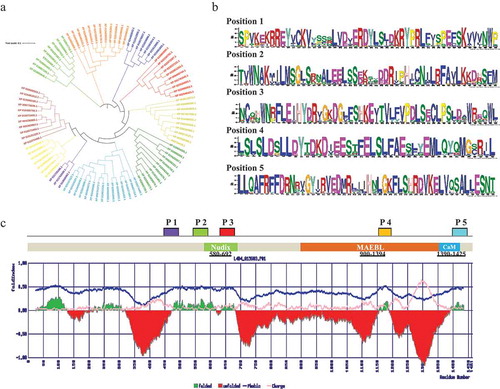
Figure 1. (a) iTOL server has been used to construct the phylogenetic tree of CCAR1 protein sequences. (b) Identification of conserved motifs of CCAR1 protein family. (c) The positions of the motifs are denoted by P1, P2, P3, P4, and P5. Conserved domain structure of M. notabilis CCAR1 protein along with predicted fold/unfold property. Despite Nudix/DBC1 homology domain, MAEBL and calmodulin (CaM) domains are also found in C-terminal of CCAR1 protein. Deleted in breast cancer domain and it homologs from diverse eukaryotes are a catalytically inactive version of the Nudix hydrolase (MutT) domain. It is predicted to bind NAD metabolites and regulate the activity of SIRT1 or related deacetylases by sensing the soluble products or substrates of the NAD-dependent deacetylation reaction.