Figures & data
Figure 1. TN content (a), SP content (b), NR activity (c) and NiR activity (d) of mulberry leaves under NaCl and NaHCO3 stress. Note: The data are from three replicated experiments (n = 3), and represent means ± SE. Significant differences were expressed by different small letters (P < .05), and very significant differences were expressed by different capital letters (P < .01).
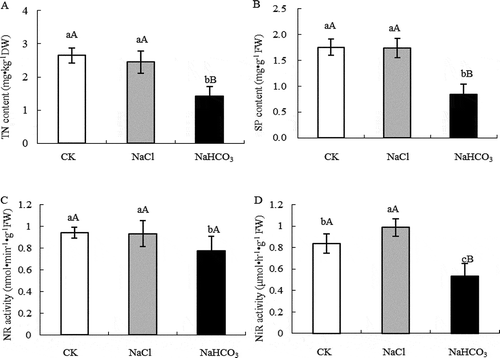
Figure 2. GOGAT activity (a) and GS activity (b) of mulberry leaves under NaCl and NaHCO3 stress. Note: The data are from three replicated experiments (n = 3), and represent means ± SE. Significant differences were expressed by different small letters (P < .05), and very significant differences were expressed by different capital letters (P < .01).
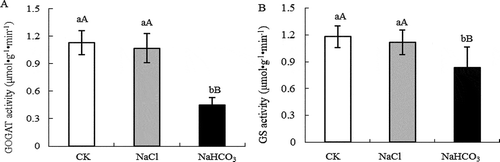
Figure 3. Pro content (a), Put content (b), Spd content (c), Spm content (d),GSH content (e) and H2O2 content (f) of mulberry leaves under NaCl and NaHCO3 stress. Note: The data are from three replicated experiments (n = 3), and represent means ± SE. Significant differences were expressed by different small letters (P < .05), and very significant differences were expressed by different capital letters (P < .01).
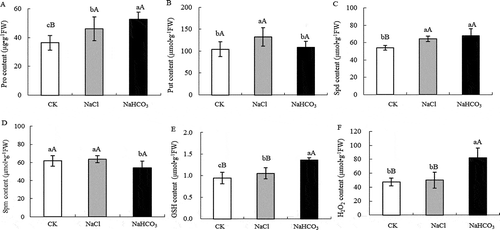
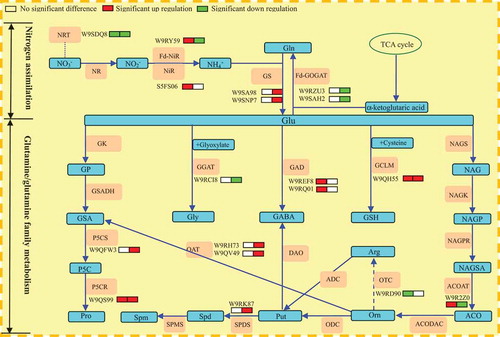
Table 1. Nitrogen assimilation and glutamine/glutamine family of amino acids metabolism related enzymes expression of mulberry leaves under NaCl and NaHCO3 stress.
Figure 4. Effects of NaCl and NaHCO3 stress on gene expression of nitrogen assimilation (a) and glutamine/glutamine family of amino acids metabolism (b) in leaves of mulberry seedlings. Note: * indicates significant difference with CK.
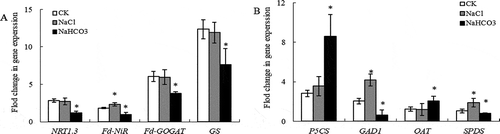
Figure 5. Schematic presentation of nitrogen assimilation and glutamine/glutamine family of amino acids metabolism of mulberry leaves under NaCl and NaHCO3 stress.