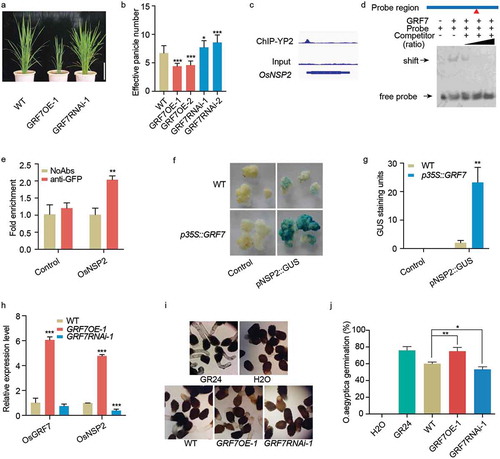
Figures & data
Figure 1. OsGRF7 controls tiller number through directly regulating strigolactone biosynthesis. (a) Plant stature of the OsGRF7 overexpression (GRF7OE-1) and RNAi (GRF7RNAi-1) transgenic plants at the tillering stage. Bar = 20 cm. (b) Effective panicle number of OsGRF7 transgenic lines. Values are means ± SD of 15 independent plants. (c) OsGRF7 binding peak in the promoter of OsNSP2. (d) EMSA validation of the binding between OsGRF7 and the promoter of OsNSP2. The two-fold, 10-fold, and 100-fold unmodified probes were used as competitors. Red triangle indicates the site of the ACRGDA motif on the promoter region. (e) ChIP-qPCR validation of the OsGRF7 binding site in the promoter of OsNSP2. The fold enrichment was normalized against the promoter of OsUBI. No addition of antibodies (NoAbs) served as a negative control. (f) OsGRF7 affects pNSP2::GUS expression in rice calli. Wild type and p35S::GRF7 calli without transfection were used as negative controls. (g) Blue pixels from the GUS staining were scanned and quantified using ImageJ (https://imagej.nih.gov/ij/). (h) Relative expression levels of the OsGRF7 and OsNSP2 detected with reverse-transcription followed by quantitative PCR (RT-qPCR). OsUBI was used as an internal reference. (i-j) Germination analysis of O. aegyptiaca seeds using the extracts of OsGRF7 transgenic lines. Distilled water (H2O) and 1 μM GR24 were used as negative and positive controls, respectively. WT, wild type. In e, g, h, and j, values are means ± SD of three biological replicates. Asterisks indicate significant difference by Student’s t-test (* P < .05; ** P < .01; and *** P < .001)