Figures & data
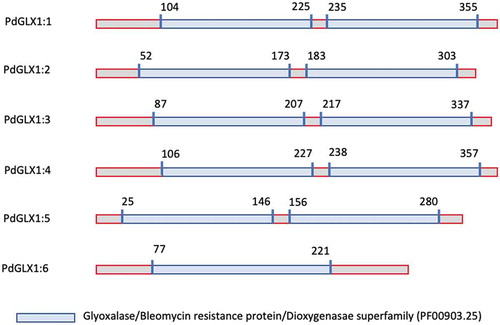
Figure 1. Phylogenetic analysis of the putative PdGLX1 proteins using the Maximum Likelihood (ML) method (a). The percentage of trees in which the proteins clustered together is shown next to the branches based on 1000 bootstrap replicates. Intron-exon map of the different PdGLX1 genes (b)
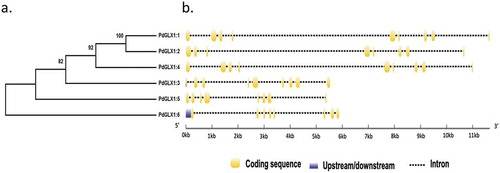
Figure 3. Multiple sequence analysis of the putative PdGLX1 proteins along with other previously well-characterized GLX1 orthologues from Oryza sativa and Arabidopsis thaliana. The Zn2+-dependent protein extended sequences are labeled as A, B, and C. Whereas, the Zn2+ (Q/E/H/E) and Ni2+ (Q/E/Q/E) metal-binding domain are shown in triangles
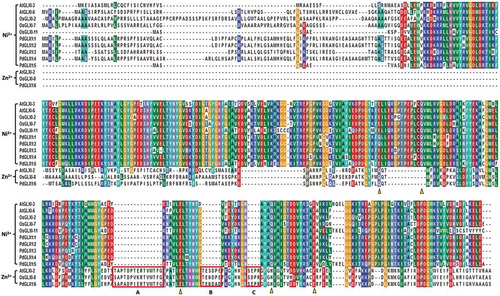
Figure 4. Conserved motifs identified by MEME of GLYOXALASE-I proteins identified from Arabidopsis thaliana, Oryza sativa, and P. dactylifera L. Lengths of motifs of each GLYOXALASE-I protein are displayed proportionally
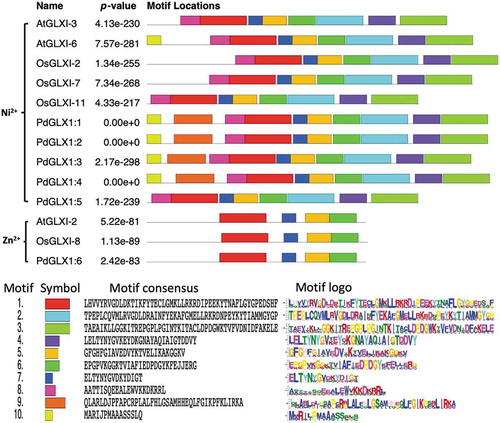
Figure 5. Phylogenetic analysis of Glyoxalase-I proteins identified from P. dactylifera L. and other plant species. Circular tree constructed for the GLXI proteins from P. dactylifera L., Arabidopsis thaliana, Oryza sativa, Sorghum bicolor, Medicago sativa, and Glycine max using Neighbor-Joining method in MEGA X with 1000 bootstrap replicates. The protein sequences are characterized as Clade I for Ni2+-dependent proteins, Clade II for Zn2+-dependent proteins, and Clade III for GLX-like proteins
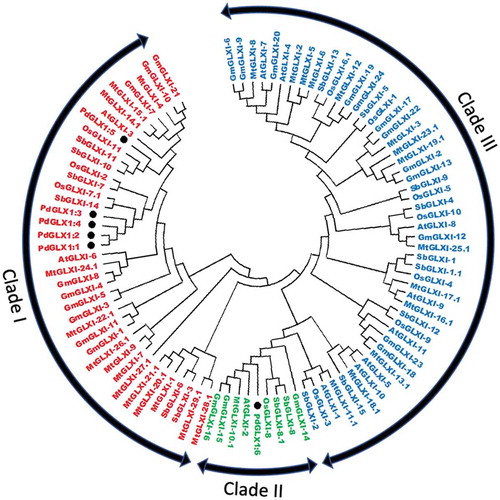
Figure 6. The relative expression level of the different PdGLX1 genes in the leaf (a) and root (b) tissue using qPCR method. Asterisk (*) indicates p 0.05; mean ±SD, n = 3
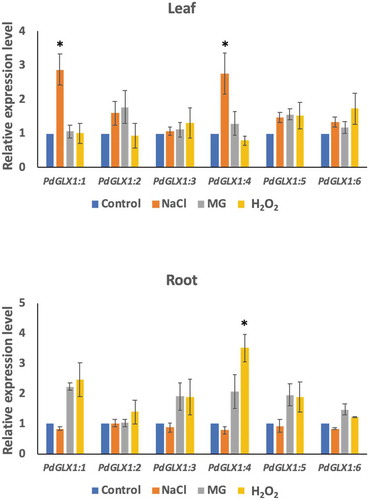
Figure 7. Heterologous expression of PdGLX1 enzymes in E. coli BL21 provides different levels of tolerance to various abiotic stresses. The PdGLX1 genes were cloned into the protein expression vector (pET-DEST52-PdGLX1:1–6) under the control of the T7 promoter (a). The transgenic bacteria were grown in the presence of different abiotic stresses, including MG (0.5 mM MG) (b), salinity (200 mM NaCl) (d), and oxidative stress (5 mM H2O2) (f). The MG concentration in the cells was measured for each treatment. The growth of the cells was spectrophotometrically monitored during the experiment. Asterisks (***) indicates p 0.001; mean ±SD, n = 3
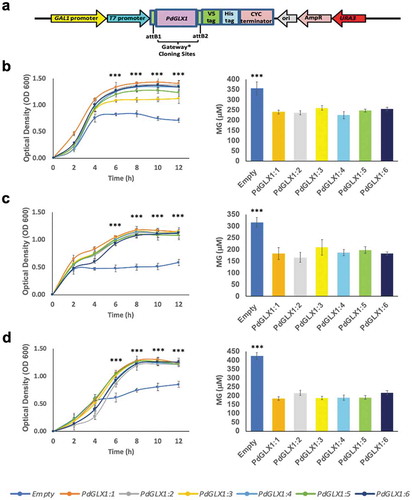
Figure 8. Mutant GLO1 yeast cells genetically transformed with the yeast expression cassette pYES-DEST52-PdGLX1:1–6 (a). The pictorial representation of the different constructs are shown (b), the transgenic cells were then streaked on Ura− SD galactose media (c), along with the different concentrations of MG; 1 mM MG (d), 2 mM MG (e), 3 mM MG (f), 4 mM MG (g), the transgenic cells were also streaked on different Ura− SD glucose media (h), along with different concentrations of MG; 1 mM MG (i), 2 mM MG (j)
Figure 9. The growth of yeast mutant cells and the estimation of their MG content. The mutant GLO1 yeast cells genetically transformed with the yeast expression cassette pYES-DEST52-PdGLX1:1–6 and the empty vector pYES-DEST52 were grown in liquid Ura− SD galactose media in the presence of 500 µM MG (a), 700 µM H2O2 (b) and 500 mM NaCl (c). Asterisks (***) indicates p 0.001; mean ±SD, n = 3
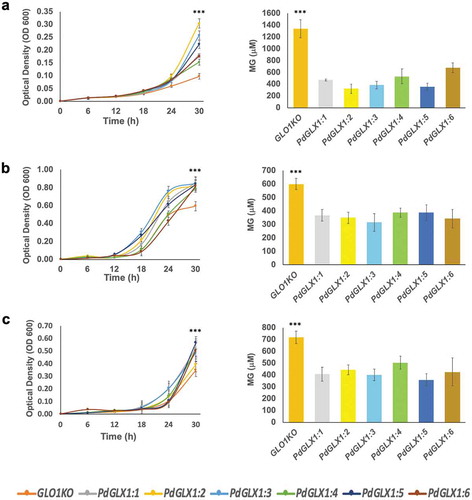
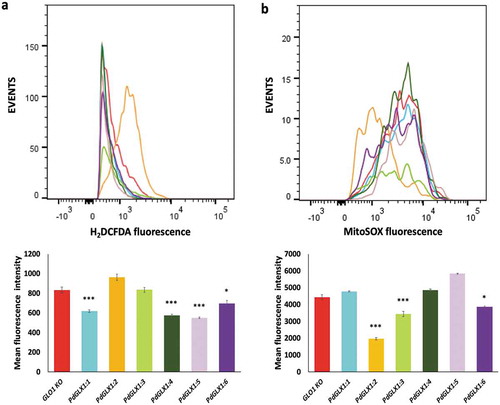
Figure 10. ROS quantification by flow cytometry in GLO1 mutant cells transformed with PdGLX1 genes or the empty negative control vector. The cells were stained to detect cytosolic ROS accumulation using H2DCFAD stain (a) or the mitochondria specific superoxide using the MitoSOX™ stain (b). Graphs indicate the mean fluorescent intensity (5000 events). Asterisks (***) indicates p 0.001, Asterisk (*) indicates p
0.05; mean ±SD, n = 3