Figures & data
Figure 1. Gene structure and amino acid sequence analysis of SbSTAR1 in sweet sorghum. (a) Gene structure of SbSTAR1 and homologous genes. Box, exon; line, intron. (b) Sequence alignment of SbSTAR1 and homologous proteins from other species, including Oryza sativa (OsSTAR1, Os06g48060.1), Arabidopsis thaliana (AtSTAR1, At1g67940.1) and Fagopyrum esculentum (FeSTAR1, AYK27446.1). Horizontal lines indicate conserved motifs of nucleotide-binding domain (NBD) in ABC transporters. (c) Phylogenic analysis of SbSTAR1 (XP_002438933.1) and its homologs in Oryza sativa, Arabidopsis thaliana, Fagopyrum esculentum, Marchantia polymorpha (PTQ46811.1), Physcomitrium patens (PNR55880.1), Chara braunii (GBG67008.1) and Escherichia coli (pstB, EGM3813836.1). The phylogenetic tree was constructed using the neighbor-joining method in MEGA 7
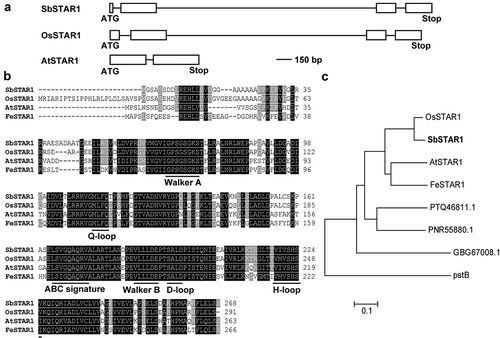
Figure 2. Quantitative real-time PCR analysis of the SbSTAR1 expression pattern. (a) Relative expression of SbSTAR1 in sweet sorghum root tips (0–1 cm) in response to 15 μM Al stress for different treatment times. (b) Relative expression of SbSTAR1 in root tips exposed to different Al concentrations for 24 h. (c) Relative expression of SbSTAR1 in root tip (0–1 cm), root (1–2 cm), root (2–3 cm) and shoot sections in the absence (-Al) or presence (+Al, 15 μM) of Al treatment for 24 h. (d) Relative expression of SbSTAR1 in root tips in response to AlCl3 (15 μM), CuCl2 (0.5 μM) and LaCl3 (10 μM) for 24 h. Data represent the means with SD (n = 3). Columns with different letters are significantly different at P < .05
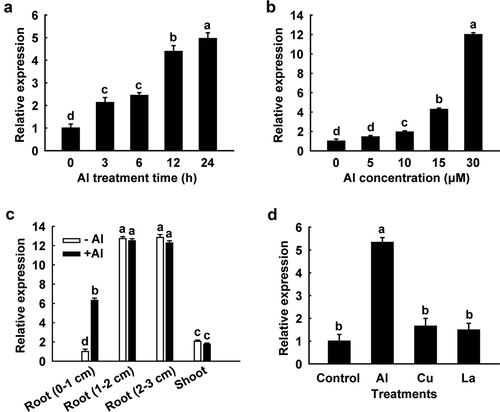
Figure 3. Transcriptional regulation of SbSTAR1 and protein subcellular localization. (a) Schematic diagram of the effector and reporter in the HEK293 expression system. In the effector, SbSTOP1 was driven by the CMV promoter; Myc, encoding Myc protein tag. In the reporter, the firefly luciferase reporter gene (Firefly Luc) was driven by the SbSTAR1 promoter (pSbSTAR1), and the Renilla luciferase reporter gene (Renilla Luc), as the internal control, was driven by the SV40 promoter. The CMV and SV40 promoter are constitutive promoters commonly used in mammalian expression vectors to drive gene expression. (b) Immunoblot analysis of the SbSTOP1-Myc fusion protein in HEK293 cells. (c) Transcriptional regulation of SbSTAR1 by SbSTOP1 in HEK293 cell. Relative luciferase activity, the luciferase activity of the reporter (Firefly Luc) was normalized to the internal control reporter (Renilla Luc). Data represent the means with SD (n = 3). ** represents significant differences from the vector-only control at P < .01. (d) Subcellular localization of SbSTAR1. Transient expression of GFP-SbSTAR1 fusion protein or GFP control in Arabidopsis protoplasts. GFP, GFP fluorescence; Bright, bright field; DAPI, nuclear signal. Scale bar indicates 20 μm
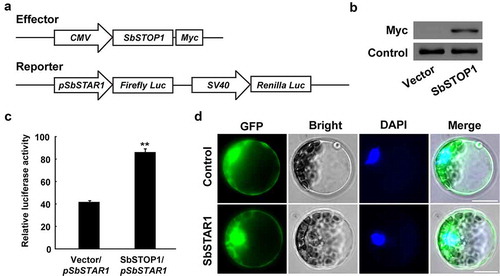
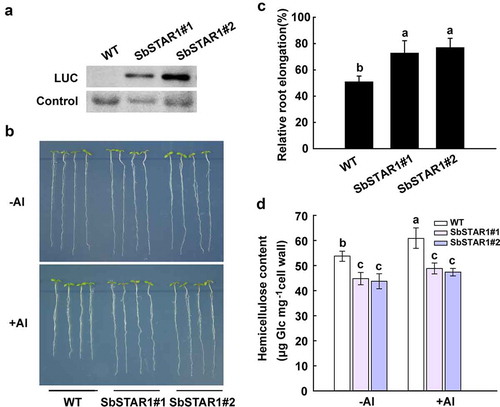
Figure 4. Transgenic Arabidopsis overexpressing SbSTAR1 shows improved tolerance to Al stress and lower hemicellulose content in root cell wall. (a) Immunoblot analysis of the LUC-SbSTAR1 fusion protein in two independent transgenic lines. (b) Root Al-sensitive phenotype of WT (Col-4) and SbSTAR1 transgenic lines. Five-day-old seedlings were cultured on MS medium and then transferred to medium containing 1 mM CaCl2 and 1% (w/v) sucrose at pH 5.0, with or without 50 μM AlCl3. (c) Relative root elongation (root elongation with Al treatment/root elongation without Al treatment) of WT and two SbSTAR1 transgenic lines. Data are shown as the means with SD (n ≥ 20). Columns with different letters indicate significant differences between plants at P < .05. Experiments were repeated three times. (d) Hemicellulose content in root cell wall of WT and two SbSTAR1 transgenic lines. The hemicellulose was extracted from root cell wall of four-week-old plants with or without 50 μM Al treatment for 24 h. Data are shown as the means with SD (n = 3). Columns with different letters indicate significant differences at P < .05