Figures & data
Figure 1. Fusarium oxysporum infection–mediated gene expression analysis. The base of a N. tabacum seedling was sampled 0, 6, 12, and 24 h after inoculation with F. oxysporum. F. oxysporum infection–mediated NtSUC4 (a), NtSTP12 (b), NtHEX6 (c), NtSWEET1 (d), NtSWEET3b (e), and NtSWEET12 (d) patterns of expression were analyzed using qRT-PCR. The mRNA levels in the samples were normalized against those of Actin mRNA. Data are the means ± SE (n = 3), and the experiments were repeated at least three times. Different letters indicate significant differences between groups (P < 0.05). qRT-PCR, Quantitative real-time reverse transcriptase–PCR; SE, standard error
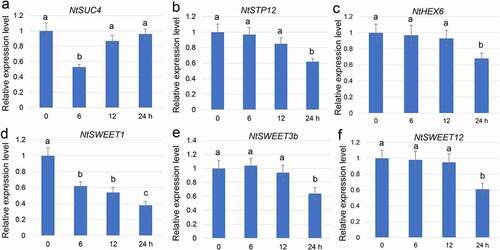
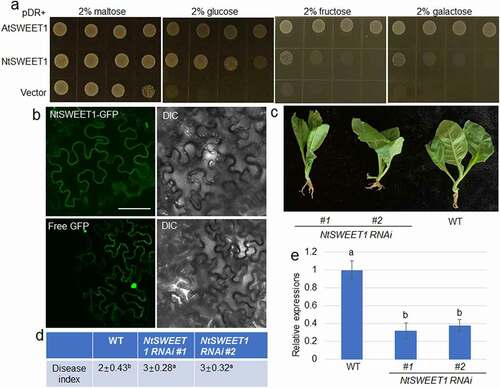
Figure 2. NtSWEET1 function and its role in defense of tobacco against root rot. (a) Hexose transport activity of NtSWEET1 was analyzed in the EBY4000 yeast strain. NtSWEET1 -transformed cells were grown on YNB media that contained 2% glucose, fructose, galactose, or maltose. Empty (pDRf1) vector and AtSWEET1 were used as the negative and positive controls, respectively. (b) Localization of NtSWEET1-GFP or free GFP was analyzed in tobacco leaves. The left and right channels indicate GFP and light image, respectively. (c) Wild-type (WT) and NtSWEET1 RNAi (#1 and #2) plants were inoculated with F. oxysporum. (d) Disease index of plants shown in (c) was calculated (n > 15). (e) Expression level of NtSWEET1 in the WT and NtSWEET1 RNAi plants (#1 and #2). Different letters indicate significant differences between groups (P < 0.05). DIC, Differential interference contrast; GFP, green fluorescent protein; WT, wild-type; YNB, yeast nitrogen base media