Figures & data
Figure 1. Effect of GIGANTEA on lateral root development in Arabidopsis seedlings. (a) Graph showing lateral root number in wild type (Col-0), 35S::GI, and gi-100 seedlings. (b) The phenotype of lateral roots in wild type (Col-0), 35S::GI, and gi-100 seedlings. All seedlings were vertically grown for 10 d on MS medium under red light, after 8 h of white light induction treatment. Averages of at least 10 seedlings ± S.D. are shown. (c) TIR1 genome track of read coverage profiles of GI in Col-0 (Orange and blue) and GI-overexpressed (green and red) lines. Peaks represent the sequence enrichment of TIR1 in Col-0 (Orange and blue) and GI-overexpressed (green and red) lines. Bioinformatic analysis of ChIP-seq data shows GI binds to the promoter region of TIR1. Read coverage is presented on the left side of each ChIP-seq. (d) Graph showing the qRT-PCR result of TIR1 gene expression relative to ACTIN in Col-0, 35S::GI, and gi-100 seedlings which are grown under red light in LD conditions. The qRT-PCR reactions were performed in triplicates. GraphPad Prism software was used to test for significance among the dataset using a one-way analysis of variance (ANOVA) followed by Sidak’s multiple comparisons test (* P < .05, ** P < .01).
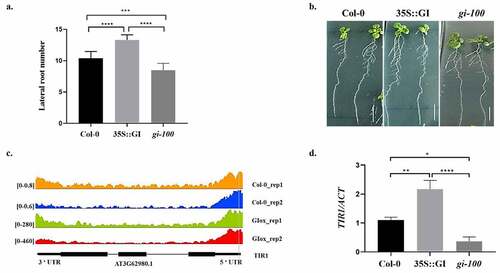
Figure 2. GIGANTEA modulates auxin signaling by regulating Aux/IAA-ARF modules in At. Relative expression of IAA14 (a), ARF7 (b), and LBD16 (c) were analyzed in wild-type (Col-0), 35S::GI, and gi-100 seedlings by qRT-PCR. All seedlings were grown for 10 d on MS medium under red light, after 8 h of white light induction treatment. Statistical analysis was performed using one-way ANOVA, with significant differences indicated relative to Col-0 (* P < .05, ** P < .01). The qRT-PCR experiments were performed in three biological replicates.
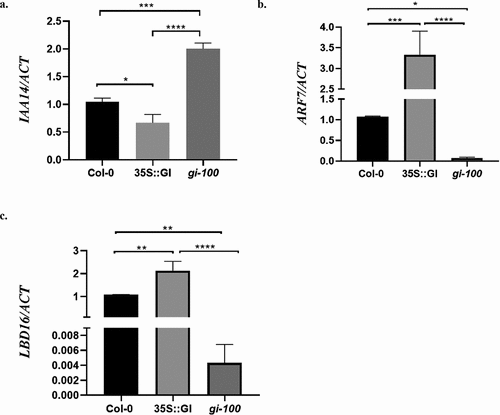
Figure 3. GI induces NAC1 and AIR3 expression in At. Transcripts abundance of NAC1(a) and AIR3 (b) were examined by qRT-PCR in wild-type (Col-0), 35S::GI, and gi-100 seedlings. Samples were harvested from 10 d old seedlings of wild type (Col-0), 35S::GI, and gi-100 grown under red light in LD conditions. One-way ANOVA was used for statistical analysis, with significant differences indicated relative to Col-0 (* P < .05, ** P < .01). Experiments were repeated three times.
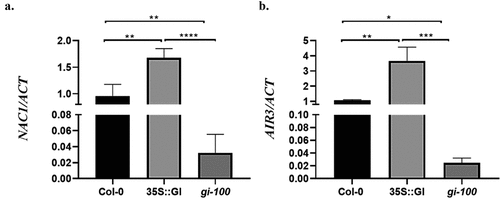
Figure 4. A Hypothetical model showing regulation of LR formation by GIGANTEA under red light in Arabidopsis. Our results suggest that in presence of red light, GI induces the density of lateral roots by modulating Aux/IAA-ARF modules. GI can bind in the promoter region of TIR1 to induce auxin signal cascade which leads to an increment in LBD16 gene transcripts during LR initiation. GI can also upregulate NAC1 expression followed by induction of AIR3 gene expression which further enhances LRs development in Arabidopsis. One probable alternate mode of regulation could be through the transcription factor, HY5. GI could make a complex with HY5 that might regulate LR formation in Arabidopsis.
Table 1. List of primers used in qRT-PCR.
