Figures & data
Figure 1. Phylogeny of class II histone deacetylase. HDA5 and HDA18 were recovered as a monophyletic group, and HDA15 in the sister clade. The sequences of animals were in a paraphyletic topology, but HDA15 of Arabidopsis and HDAC of humans belong to a sister clade suggesting functional conservation; in green HDAC class II of Arabidopsis and red HDAC class II of Homo sapiens. Soybean (SOYBN), Oriza sativa japonica (ORYSJ).
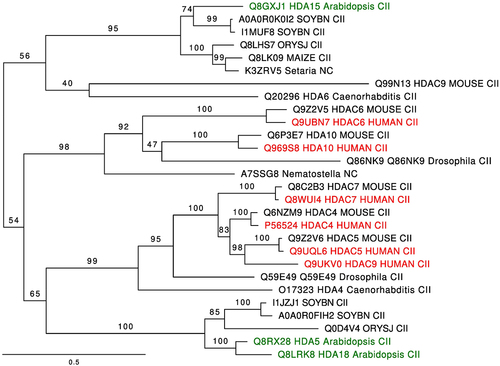
Figure 2. MADS-domain/MEF2 motif is conserved in some MADS-domain proteins of Arabidopsis. (a) Graphic representation of MEF2 protein, in red the MADS-domain and in orange the MEF2-domain. Below the model representation, there is the linear amino acid sequence indicating the MADS-domain, HDAC binding region and MEF2 domain (Figure was modified from Lu et al. 2000Citation40). (b) Multiple alignments of the MEF2C motif sequence, tomato MADS-domain proteins, and some Arabidopsis MADS-domain proteins. In orange are identical amino acids, in yellow less conserved substitutions, and in blue conservative substitutions. Homo sapiens (Hs), Lycopersicon esculentum (Le), Arabidopsis thaliana (At).
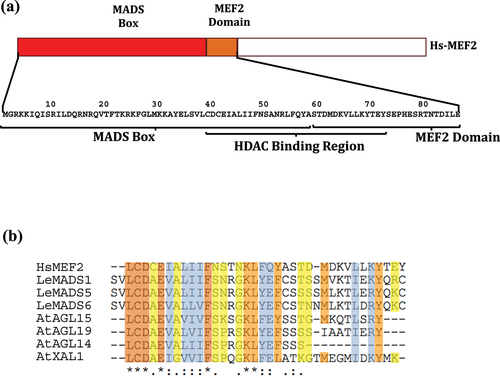
Table 1. Interaction of XAL1 with HDA15 in Arabidopsis as predicted by HDOCK docking simulations. The table shows the top ten models that docking analysis generated and the reported interaction for humans of MEF2B with HDAC9. The two last rows show the images of the structure of the interactions. HDA15 is in light lilac, XAL1 is in helices green and aquamarine, and the last column is HDAC9 and MEF2B in aquamarine and green, respectively.
Figure 3. Expression of MADS and HDACs in root of Arabidopsis thaliana. XAL1 expression HDACs and different times in the development of the root from Arabidopsis (PCR endpoint); days after sowing (DAS), tubulins (TUB).
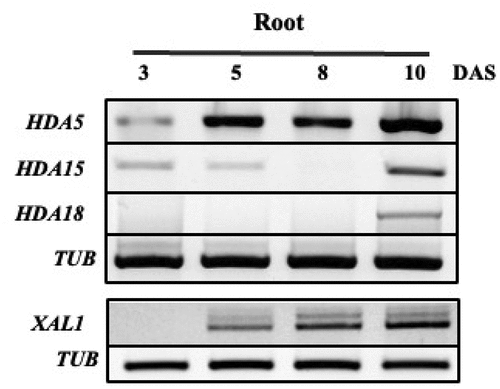
Figure 4. Protein-protein interaction between HDA15 and XAL1. A two-hybrid assay indicates an interaction between HDAC and XAL1 in Arabidopsis. (a) SD/-L/-W/-H correspond to dropout medium lacking L, W, H and positive interactions, BD HDA15 and AD XAL1, resulting in yeast growth on the SD/-L/-W/-H plate. (b) HDA15 and XAL1 interaction with different concentrations of 3-AT (0.25, 0.5, 0.75, 1.0, 1.25 and 1.5 mM). (c) HDA15 interaction with XAL1 at different dilutions (1:50 and 1: 1000). (d) Lac-Z expression in interaction HDA15 and XAL1. AGAMOUS LIKE 6 (AGL6) and APETALA 3 (AP3) was used as a positive control.
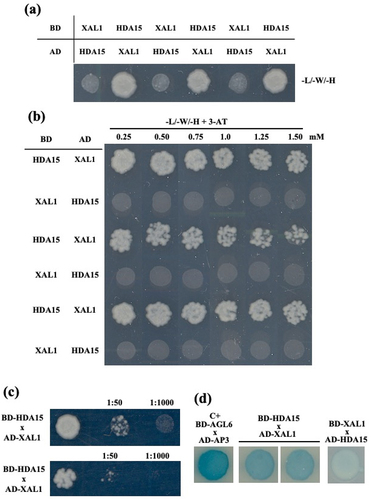
Figure 5. Subcellular localization of the interaction of HDA15 and XAL1. Bimolecular fluorescence complementation assay in tobacco leaf cell nuclei between transiently expressed HDA15 and XAL1. (a) HDA15-XAL1 interaction in the nucleus (YFC-XAL1 + YFN-HDA15). (b) Positive control, PISTILLATA interaction with APETALA3 (YFC-PI + YFN-AP3). In c and d is a zoom of a and b, respectively. In c, the yellow arrow points to the possible nucleolus. The GFP spectrum is shown in the left column panels. Merged visible and fluorescent signals are shown in the right column panel. Scale bar: 20 m.
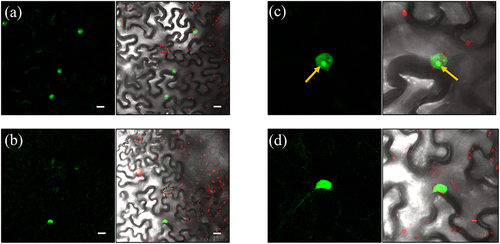
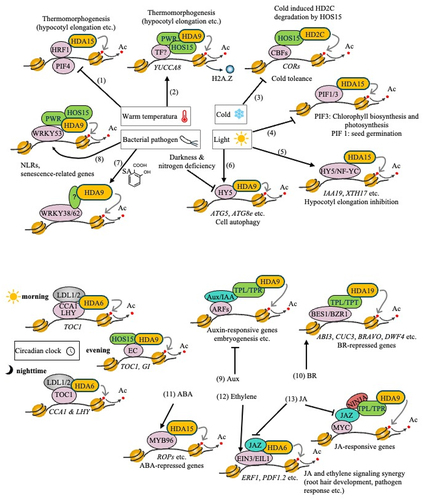
Figure 6. Summary of principal function of histone deacetylation in plants. 1, repression thermal-responsive genes and warm temperatures induce PIF4 inhibiting HDA15/HFR1 interaction. 2, hypoacetylation at the YUCCA8 by HDA9/PWR/HOS15 complex in warm temperatures. 3, HOS15/CBFs/HD2C complex repress COR genes at normal temperature; the cold induces HD2C degradation. 4, HDA15, PIF1, and PIF3 are associated in dark conditions, deacetylating histones (3 and 4) to repress chlorophyll biosynthetic/photosynthetic and seed germination genes. In light, HDA15 is released to degradation of PIF1 and PIF3. 5, in light, the HDA15-NFYCs-HY5 complex inhibits the expression of IAA19 and XTH17 genes, which are related to auxin biosynthetic, signaling, and cell wall organization. 6, in light, the HY5/HDA9 interaction inhibits ATG5 and ATG8e (cell autophagy genes). In Darkness and nitrogen deficiency, these genes are released for the degradation of HY5-inducing autophagy. In bacterial infection: 7, SA induction occurs, WRKY38/62 is expressed, and WRKY38/62 recruits HDA19 to elicit a basal defense response.; 8, the expression of WRKY53 is induced, recruiting the HDA9/PWR/HOS15 complex. This complex suppresses NLR genes and leaf senescence genes. Hormone signaling: 9, HDA19 and Aux/IAA – TPL/TPR induced deacetylation, and auxin-responsive genes were repressed without Aux. In the presence of Aux, the Aux/IAA protein is degraded, and ARFs are released; 10, BR stimulates BES1 and BZR1 activity to recruit TPL/TPR/HDA19 complex and repress BR-repressed genes; 11, High concentrations of ABA stimulate the MYB96 - HDA15 interaction, regulating ABA-repressed genes for hyperacetylation of histones (3 and 4); 12, JAZ and HDA6 repress transcription factors EIN3/EIL1, and ethylene stabilizes EIN3/EIL1. JAZ is degraded for JA to express ethylene-responsive genes; 13, the interaction of JAZ and NINJA with TPL/TPR recruits HDA19 and HDA6 for transcriptional repression in the absence of JA. Circadian clock: 14 morning, HDA6/LDL1–2/CCA1/LHY complex repress TOC1; 14 evening, Evening Complex (EC), HDA9 and HOS15 repress TOC1 and GI expression; 14 nighttime, HDA6, LDL1/2, TOC1 repress the expression of CCA1/LHY. ABA INSENSITIVE3 (ABI3). AUTOPHAGY-RELATED GENES (ATG5/8). BRASSINOSTEROIDS at VASCULAR and ORGANIZING CENTER (BRAVO). CIRCADIAN CLOCK-ASSOCIATED1 (CCA1). COLD-RESPONSIVE (COR). CUP SHAPED COTYLEDON3 (CUC3). DWARF4 (DWF4). ETHYLENE RESPONSE FACTOR1 (ERF1). INDOLE-3-ACETIC ACID INDUCIBLE 19 (IAA19). LATE ELONGATED HYPOCOTYL (LHY). PLANT DEFENSIN 1.2 (PDF1.2). RHO of PLANTS (ROP). XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17 (XTH17). TIMING of CAB EXPRESSION 1 (TOC1). Auxin (Aux). Abscisic Acid (ABA). Salicylic Acid (SA). AUXIN RESPONSE FACTOR (ARFs). Brassinosteroid (BR). BRI1-EMS-SUPPRESSOR1 (BES1). BRASSINAZOLE-RESISTANT1 (BZR1). C-REPEAT BINDING FACTORSs (CBFs). CIRCADIAN CLOCK-ASSOCIATED1 (CCA1). ELONGATED HYPOCOTYL5 (HY5). ETHYLENE INSENSITIVE 3 (EIN3). ETHYLENE INSENSITIVE 3-LIKE 1 (EIL1). HIGH EXPRESSION of OSMOTICALLY RESPONSIVE GENES15 (HOS15). HD2-type HDACs (HD2C). VARIANT HISTONE 2A (H2A.Z). INDOLE-3-ACETIC ACID (IAA). JASMONATE ZIM DOMAIN (JAZ). Jasmonic Acid (JA). LONG HYPOCOTYL in FAR-RED1 (HRF1). LSD1-LIKE1/2 (LDL1/2). LYSINE-SPECIFIC DEMETHYLASE1 (LSD). LATE ELONGATED HYPOCOTYL (LHY). TRANSCRIPTION FACTOR MYB 96 (MYB96). TRANSCRIPTION FACTOR MYC (MYC). NOVEL INTERACTOR of JAZ (NINJA). NUCLEAR FACTOR-YC HOMOLOGS (NF-YCs). PHYTOCROME-INTERACTING FACTOR (PIF). POWERDRESS (PWR). TRANSCRIPTION FACTOR (TF). TOPLESS (TPL). TOPLESS- RELATED (TPR). TIMING of CAB EXPRESSION (TOC). TRANSCRIPTION FACTOR WRKY (WRKY). Figure was modified Citation53, Citation54.