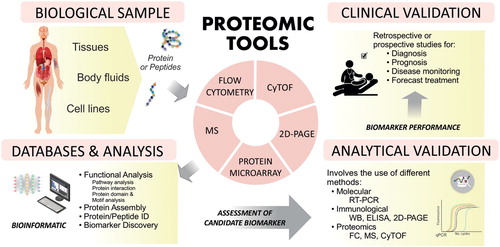
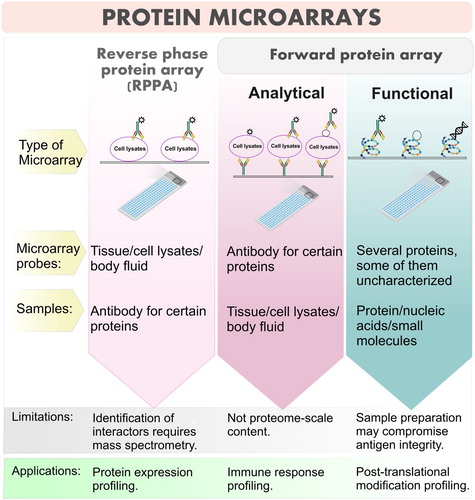
Figures & data
Table 1. Current markers with the clinical application for the classification of BCP-ALL and monitoring of BCP-ALL MRD.
Table 2. Examples of potential molecular biomarkers for diagnosis, prognosis, and targeted therapy of BCP-ALL.
Table 3. Proteomic tools in studies of BCP-ALL.