Figures & data
Figure 1. Heat maps of potential cell types in PMF and control groups, (a): cell types of GSE61629 dataset; cell types of GSE 26049 dataset; (b): the corporate cell types in the GSE61629 and GSE26049 datasets.(c): volcano plot of DEGs in GSE61629 dataset; (d): volcano plot of DEGs in GSE26049 datasets; (e): heat map of the top 40 DEGs in GSE61629 dataset; (f): heat map of the top 40 DEGs in GSE26049 dataset.
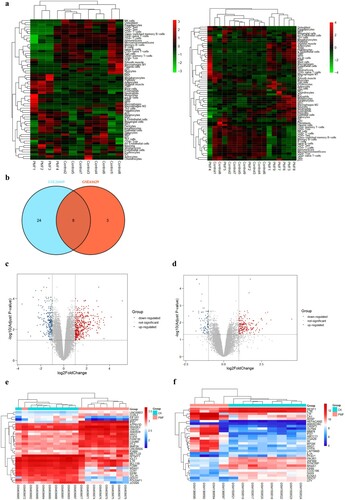
Figure 2. Functional and pathway enrichment analyses of DEGs, (a): Venn diagram displays the common DEGs in the GSE61629 and GSE26049 datasets; (b): the enrich circle plot presents the top 20 most remarkable enriched GO terms, different color module represents various term; (c): the bubble plot reflects the result of KEGG pathway analysis.
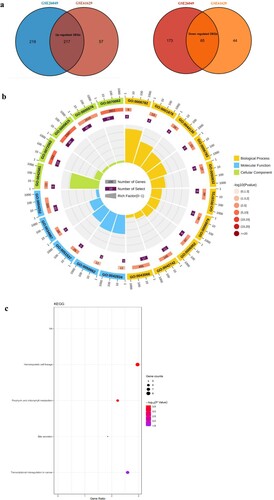
Figure 3. PPI network and hub gene selection, (a): PPI network of 282 DEGs; (b): five topological analysis methods based on CytoHubba; (c): Venn plot reflects the intersected hub genes based on five taxonomic methods.
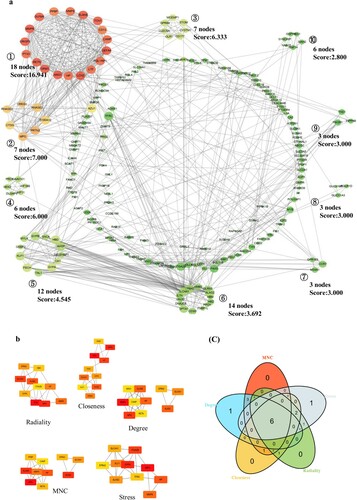
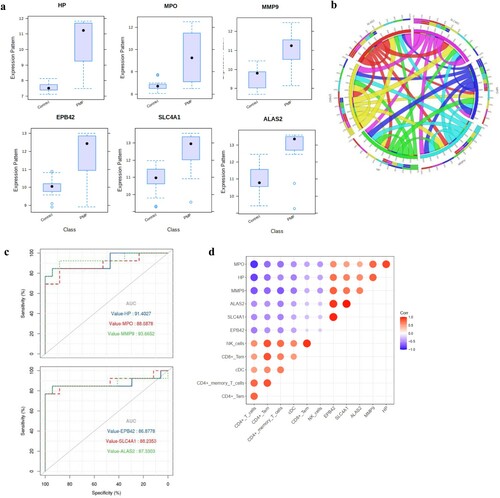
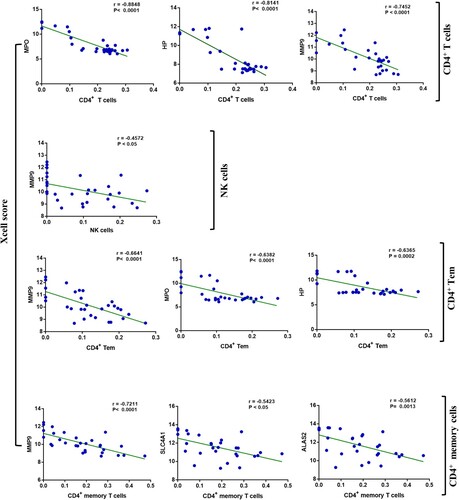
Figure 4. Characteristics of six hub genes, (a): the expression of each hub gene in the two datasets; (b): the correlation chordal graph of hub genes; (c): ROC curve of each hub genes; (d): the correlation heat map of hub genes and significant immune cells.