Figures & data
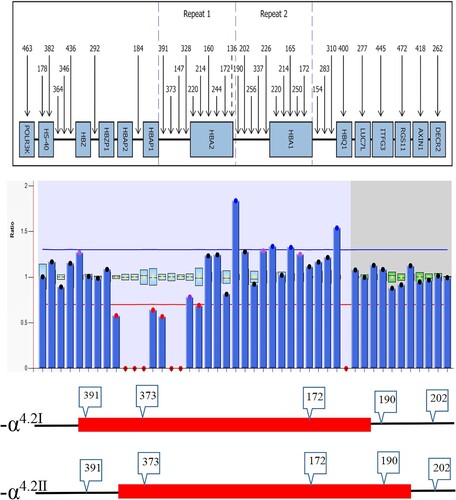
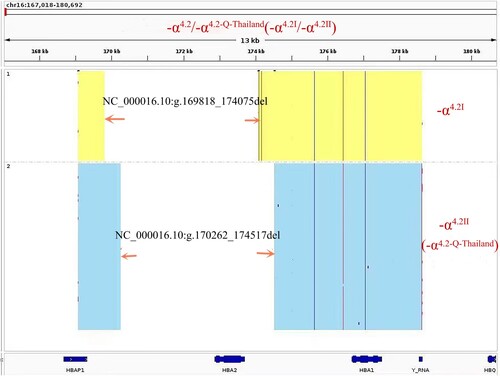
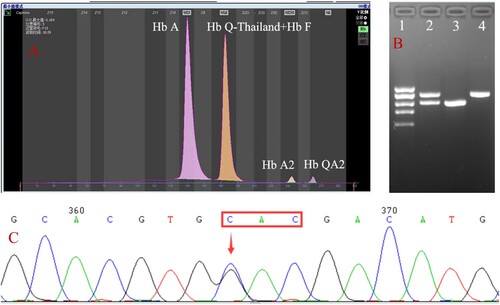
Figure 1. Hb analysis by CE showed an abnormal peak migrating to zone F (A). Results of agarose electrophoresis of Gap-PCR amplification products (B): 1. DL2000 (from top to bottom: 2.0, 1.7, 1.4, 1.2, 0.9 kb), 2. -α4.2/αα, 3. -α4.2/-α4.2, 4. Normal. A mutation of GAC>CAC in a heterozygous state was observed at codon 74 of the HBA1 gene using Sanger sequencing, corresponding to Hb Q-Thailand (C).
Data availability statement
All data in this study are shown in the figures and tables.