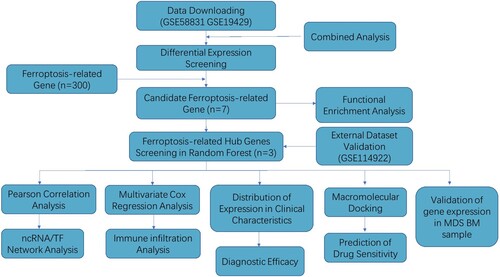
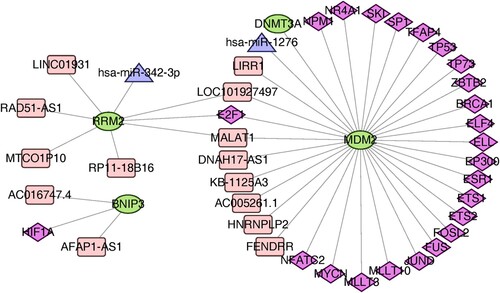
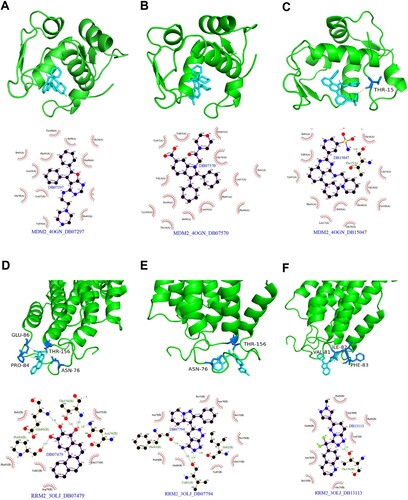
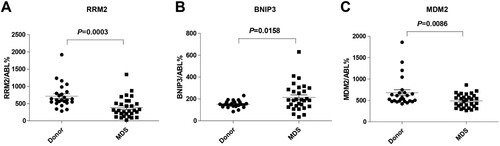
Figures & data
Figure 2. Identification of Ferroptosis-Related DEGs between MDS and healthy control. A. Volcano plot of genes differentially expressed between MDS patients and health in the dataset GSE58831. Green nodes represent down-regulation in MDS; red nodes represent up-regulation; and gray nodes represent no significant difference from healthy. B. Statistics on the results of volcano plots. C. Venn diagram to identify ferroptosis-related DEGs between MDS patients and healthy; D. The expression trend of 50 overlapping genes in MDS patients. E. Venn diagram to identify ferroptosis-related DEGs between the GSE58831 dataset and the GSE19429 dataset.
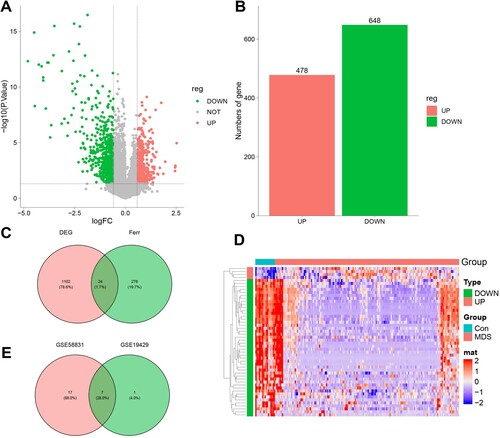
Figure 3. Enrichment analysis of 7 Ferroptosis-Related DEGs. A. 7 Ferroptosis-Related DEGs GO biological processes pathway. B. Ferroptosis-Related DEGs GO molecular function pathway. C. Ferroptosis-Related DEGs cellular component pathway. D. Ferroptosis-Related DEGs KEGG pathway.
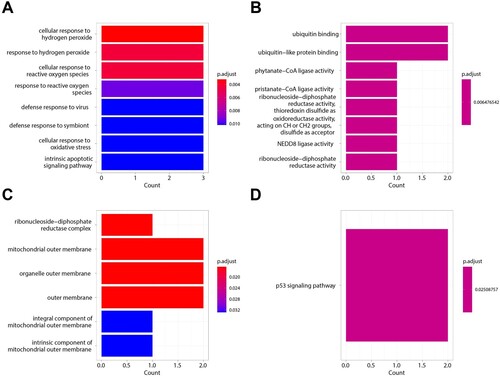
Figure 4. Ferroptosis-related Genes screening. A. The relationship between the accuracy of the random forest model and mtry in the GSE58831 dataset. B. The relationship between the error rate of the random forest model and tree in the GSE58831 dataset. C. The MeanDecreaseAccuracy of Ferroptosis-related Genes in the GSE58831 dataset. D. The MeanDecreaseGini of Ferroptosis-related Genes in the GSE58831 dataset. E. Venn diagram to identify ferroptosis-related DEGs between the GSE58831 dataset and the GSE19429 dataset. F. The correlation between MDM2, RRM2, and BNIP3 by Pearson correlation coefficient in the GSE58831 dataset.
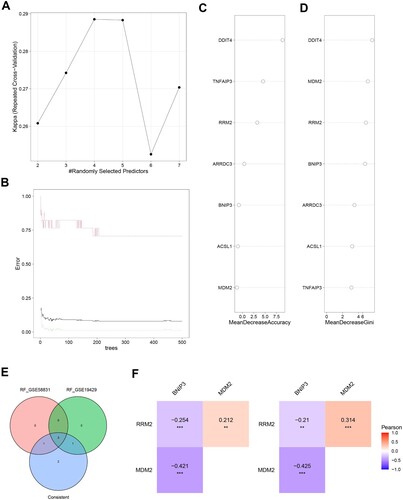
Figure 6. Classification of MDS Based on the Ferroptosis-Related Gene Sets. A. The expression of Ferroptosis-Related Gene in MDS patients and healthy in the GSE58831 dataset. B. The expression of BINP3, MDM2, and RRM2 in MDS patients with different cytogenetics in the GSE58831 dataset. C. The expression of three hub genes in MDS patients with different IPSS risk scores in the GSE58831 dataset. D. The expression of hub genes in MDS patients with different WPSS risk scores in the GSE58831 dataset. E. The expression of Ferroptosis-Related Gene in MDS patients and healthy in the GSE19429 dataset. F. The expression of hub genes in MDS patients with different WHO subtypes in the GSE19429 dataset.
Figure 7. Ferroptosis-related Genes screening and diagnostic efficacy test and Evaluation of predictive efficacy of prognostic models. A. ROC curve showing the diagnostic efficacy of BNIP3, MDM2, and RRM2 in the GSE58831 dataset. B. ROC curve showing the diagnostic efficacy of BNIP3, MDM2, and RRM2 in the GSE19429 dataset. The horizontal axis represented the specificity of the false positive rate, and the vertical axis represented the sensitivity of the true positive rate. C. KM survival curves of two risk score groups in the GSE58831 dataset. D. ROC curves in the GSE58831 dataset and the 1-year overall survival rates were 0.7.
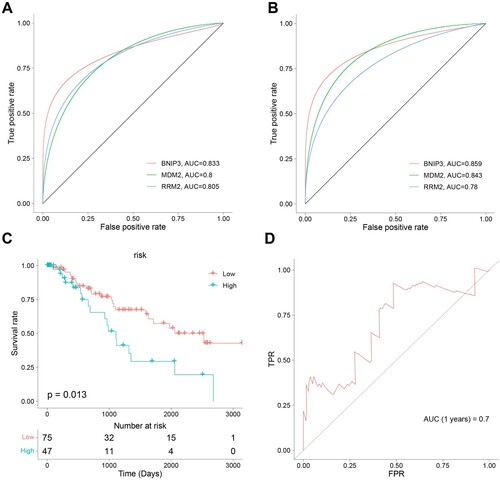
Figure 8. Correlation between the hub genes signature of BNIP3, MDM2, RRM2, and immune microenvironment. A. Correlation between risk genes and different immune infiltration cell types. B. The correlation of risk genes BNIP3, MDM2, and RRM2 expression with immune infiltration cell types in the high- and low-risk groups. C-F. The correlation of risk score with CD8+ T cells, naïve T cells CD4, regulatory T cells (Tregs), and activated myeloid dendritic cells.
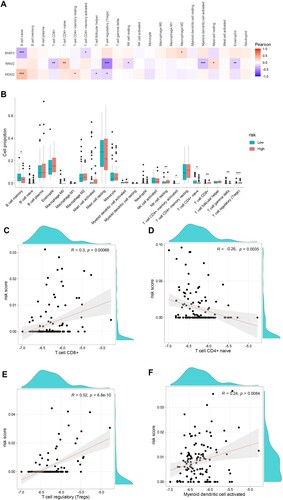
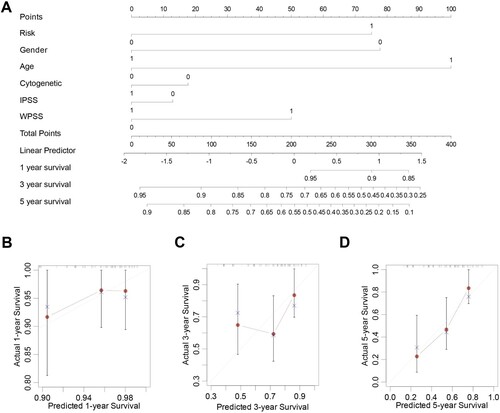
Figure 9. Nomogram construction. A. Nomogram predicting 1-, 3-, and 5-year OS for the MDS patients. (B-D). Calibration curve for the nomogram.
supple_Fig1.jpg
Download JPEG Image (363.3 KB)supple_Fig4.jpg
Download JPEG Image (201.1 KB)supple_Fig3.jpg
Download JPEG Image (209 KB)Data availability statement
Publicly available datasets were analyzed in this study and can be obtained from GEO (https://www.ncbi.nlm.nih.gov/gds/).