Figures & data
Table 1. Molecular modelling data for compounds 4, 8, 9, 16 and S-58 during docking in the active site of the COX-2 enzyme (PDB: ID 1CX2).
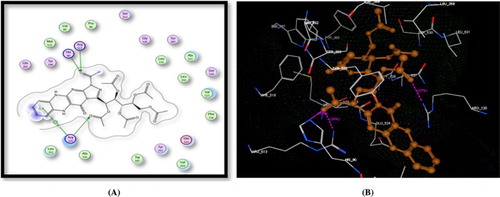
Figure 1. A) Binding of the candidate 4 with COX-2 (using MOE site finder programme), the dotted lines represent H-bonding interactions between oxadiazole N and His90 and between the acetyl C = O atom and Tyr355& Arg120. B) 3D interactions of 4 with Tyr355, Arg120, and His90 acid residues.
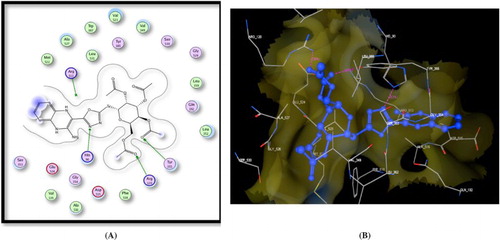
Figure 2. A) Binding of the candidate 8 with COX-2 (using MOE site finder programme), the dotted lines represent H-bonding interactions between NH and Leu352, pteridine C = O, and Gln292 and between the acetyl C = O atom and Arg513. B) 3D interactions of 8 with Leu352, Gln292, and Arg513 acid residues.
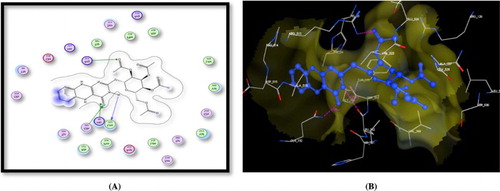
Figure 3. A) Binding of the candidate 9 with COX-2 (using MOE site finder programme), the dotted lines represent H-bonding interactions between acetyl C = O and Ser530 & Tyr385 and pteridine C = O and Arg513 and the arene cation interactions between the phenyl ring and Arg513 & Arg120. B) 3D interactions of 9 with Ser530 & Tyr385 and Arg513 acid residues.
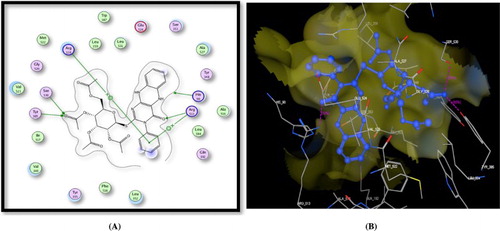
Figure 4. A) Binding of the candidate 16 with COX-2 (using MOE site finder programme), the dotted lines represent H-bonding interactions between acetyl C = O and His90 & Arg513 and Arg120 and the arene cation interactions between the benzene ring and Arg120. B) 3D interactions of 16 with His90 & Arg513 and Arg120 acid residues.

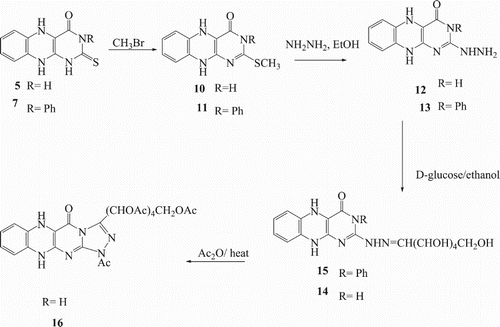
![Scheme 2. Synthesis of O-acetyl-β-D-glucopyranosyl-thioxobenzo[g]pteridin-derivatives 8 and 9.](/cms/asset/b509c732-a3e3-4191-86d7-ea00635864ea/tusc_a_1510163_f0006_c.jpg)