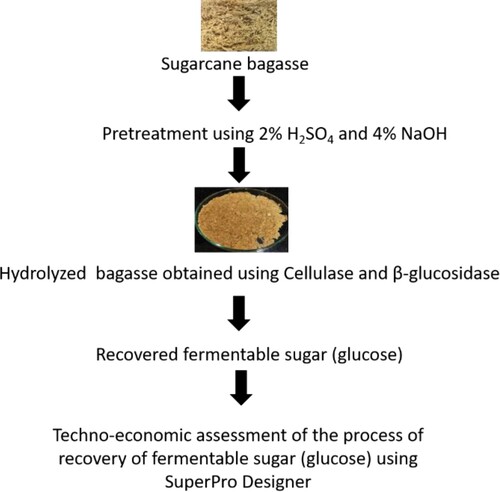
Figures & data
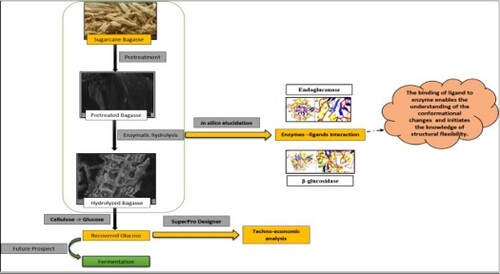
Figure 2. Structural morphology of sugarcane bagasse: (a) before hydrolysis, (b) after pretreatment and (c) after hydrolysis.
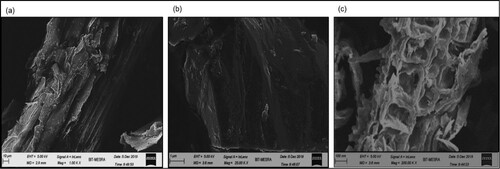
Figure 4. (a) TGA curve (b) DTA curve of sugarcane bagasse [BP – before pretreatment; AP – after pretreatment; AH – after hydrolysis].
![Figure 4. (a) TGA curve (b) DTA curve of sugarcane bagasse [BP – before pretreatment; AP – after pretreatment; AH – after hydrolysis].](/cms/asset/f05a3b4d-c23a-4074-b554-69e88abf06cb/tusc_a_2040243_f0004_oc.jpg)
Table 1. Elemental analysis of sugarcane bagasse.
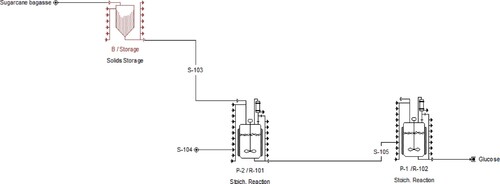
Figure 6. Cost analysis of the process obtained using SuperPro Design: (a) overall cost and (b) item cost.
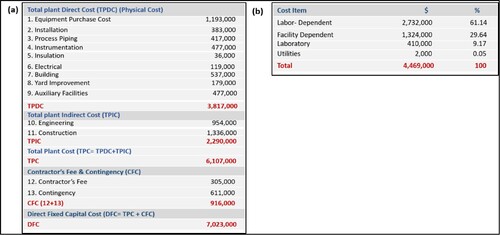
Figure 7. Cartoon representation of PBD structure: (i) Generated model viewed in Chimera (a) Endoglucanase and (b) β-glucosidase, (ii) Ligands (a) Cellulose and (b) Glucose [Yellow: Hydrogen, Blue: Oxygen, Red: Carbon], (iii) Endoglucanase (a) Chain A of 3QX3 structure for cluster 1 after docking of a molecule with the ligand using Swiss Dock and (b) The magnified image of the interaction of molecule with ligand; (iv) Beta-glucosidase (a) Chain A of 3WBE structure for cluster 5 after docking of a molecule with the ligand using Swiss Dock and (b) The magnified image of the interaction of molecule with ligand [Red colour: Coil, Yellow colour: Helix, Blue colour: Strand, Cyan colour: Ligand] obtained by Swiss Dock after docking.
![Figure 7. Cartoon representation of PBD structure: (i) Generated model viewed in Chimera (a) Endoglucanase and (b) β-glucosidase, (ii) Ligands (a) Cellulose and (b) Glucose [Yellow: Hydrogen, Blue: Oxygen, Red: Carbon], (iii) Endoglucanase (a) Chain A of 3QX3 structure for cluster 1 after docking of a molecule with the ligand using Swiss Dock and (b) The magnified image of the interaction of molecule with ligand; (iv) Beta-glucosidase (a) Chain A of 3WBE structure for cluster 5 after docking of a molecule with the ligand using Swiss Dock and (b) The magnified image of the interaction of molecule with ligand [Red colour: Coil, Yellow colour: Helix, Blue colour: Strand, Cyan colour: Ligand] obtained by Swiss Dock after docking.](/cms/asset/0ef248d4-084d-409f-83fe-6fcd61fe3095/tusc_a_2040243_f0007_oc.jpg)
Table 2. Summary of successfully produced models for endoglucanase and β-glucosidase.
Table 3. Plot statistics of the generated models using PROCHECK [A: Endoglucanase III and B: β-glucosidase].
Table 4. Poses obtained through molecular modelling using Swiss Dock.