Figures & data
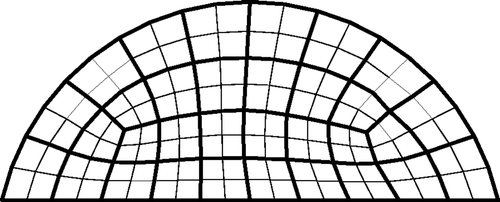
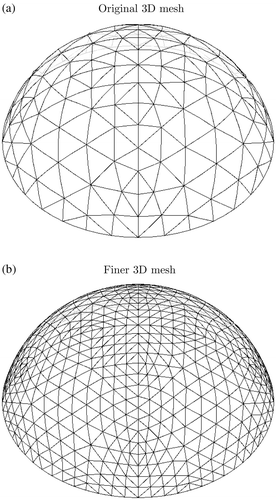
Figure 2. Finite element meshes used to represent individuals (a) Two-dimensional mesh and (b) Three-dimensional mesh.
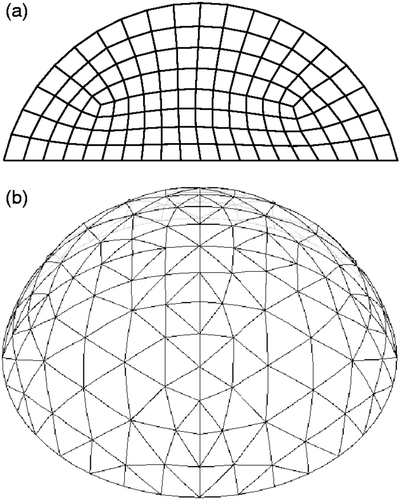
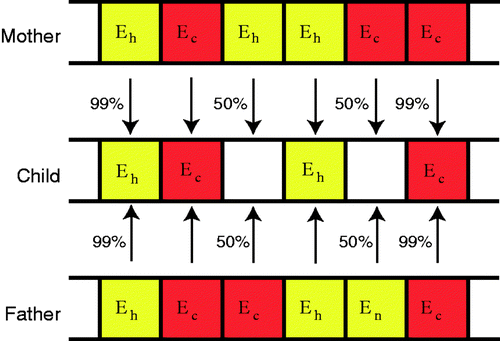
Figure 3. Chromosome used to encode material property information for each individual. (Eh is healthy modulus, Ec is cancerous modulus.)
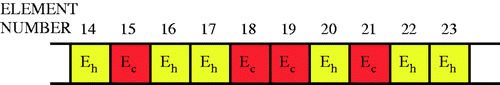
Figure 6. Challenging test case for 20 trials with SNR = 23 dB. Bold/green/grey lines show the location of the test tumours, and the numbers in the elements indicate how many times the algorithm identified that clump of elements as a tumour. For every trial at least eight elements were identified as cancerous.
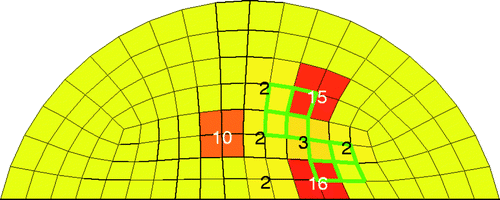
Figure 7. Results for eight different test cases with SNR = 23 dB and 20 trials. Bold/green/grey lines show the location of the test tumours, and the numbers in the elements indicate how many times the algorithm identified that clump of elements as a tumour. For every trial containing a test tumour at least four elements were identified as cancerous.
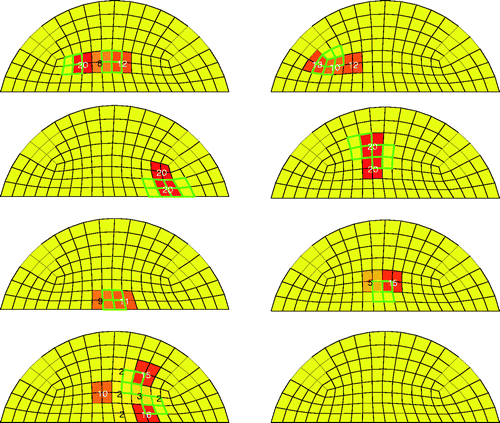
Figure 8. Typical result (Test A) at SNR = 23 dB. The true tumour crosses two clumps, adjacent to the chest wall and tissue centerline. True tumour is shown in bold/green/grey and prediction is red/black. True tumour volume is 0.96 cm3. (a) Bottom view and (b) Side view.
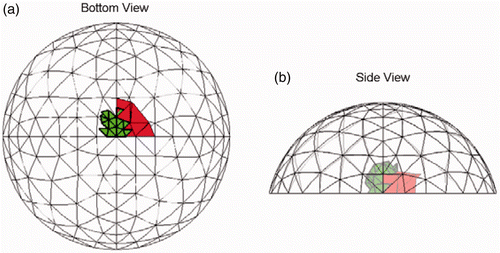
Figure 9. Result for Test B (20/20 trials) at SNR = 23 dB. The true tumour crosses eight clumps. True tumour is shown in bold/green/grey and prediction is red/black. True tumour volume is 1.02 cm3. (a) Bottom view and (b) Side view.
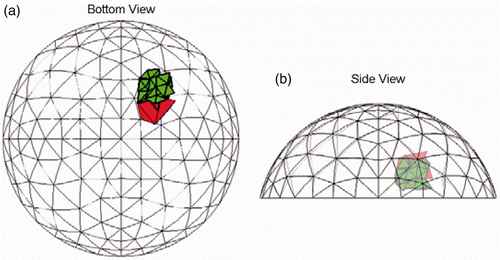
Figure 10. Result for Test C (20/20trials) at SNR = 23 dB. The true tumour crosses eight clumps. True tumour is shown in bold/green/grey and prediction is red/black. True tumour volume is 1.13 cm3. (a) Bottom view and (b) Side view.
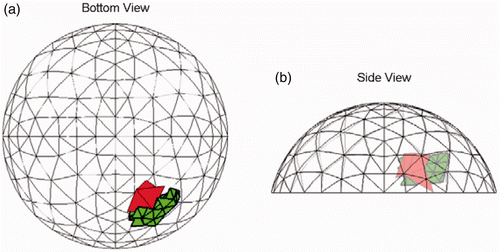
Figure 11. Most common (9/20 trials) result for Test D at SNR = 23 dB. The true tumour crosses four clumps. True tumour is shown in bold/green/grey and prediction is red/black. True tumour volume is 0.99 cm3. (a) Bottom view and (b) Side view.
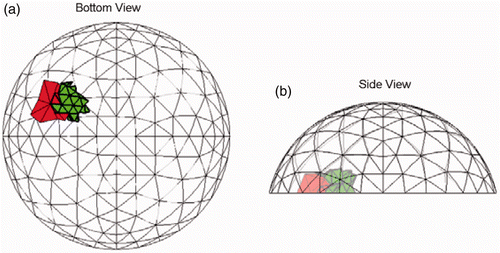
Figure 12. Most common (16/20 trials) result for Test E at SNR = 23 dB. Each of the sites in the true tumour crosses eight clumps. True tumour is shown in bold/green/grey and prediction is red/black. True tumour volume is 1.92 cm3, divided roughly equally between the two sites. (a) Bottom view and (b) Side view.
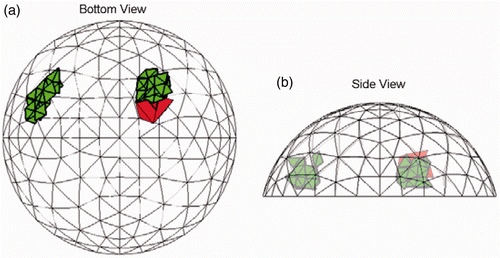
Figure 13. Result for Test F (20/20 trials) at SNR = 23 dB. The true tumour crosses eight clumps. True tumour is shown in bold/green/grey and prediction is red/black. True tumour volume is 2.07 cm3. (a) Bottom view and (b) Side view.
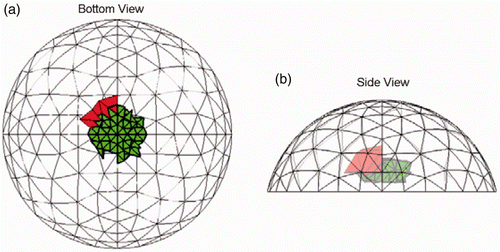
Figure 14. Result for Test G (20/20 trials) at SNR = 23 dB. The true tumour crosses twenty-two clumps. True tumour is shown in bold/green/grey and prediction is red/black. True tumour volume is 2.10 cm3. (a) Bottom view and (b) Side view.
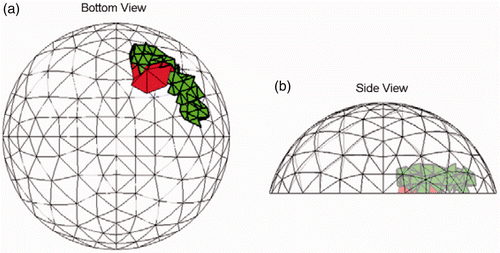
Table 1. Parameter settings for 2D and 3D GA.
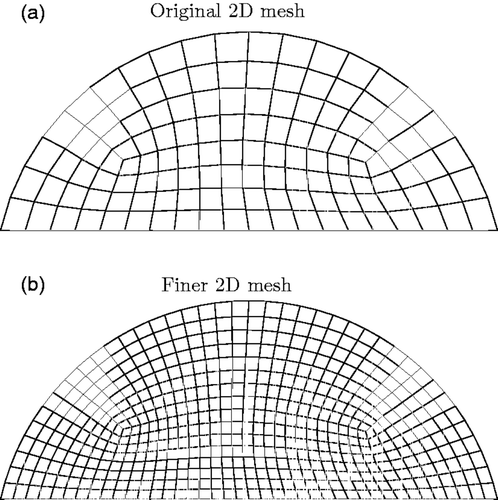
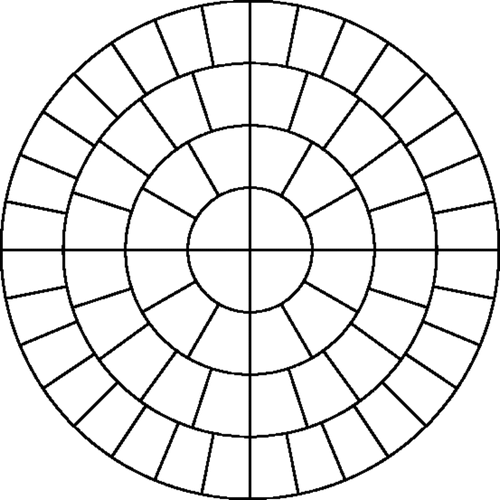
Figure D1. Finer mesh used to create measured data for original mesh in two dimensions. (a) Original 2D mesh and (b) Finer 2D mesh.