Figures & data
Figure 1. Predictions for the year 2050 on the number of deaths caused by various diseases, including antimicrobial-resistant diseases. (From: review on antimicrobial resistance. Tackling drug-resistant infections globally: final report and recommendations. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 27 December 2019)) (with permission of Wellcome collection, Attribution 4.0 International (CC BY 4.0)).
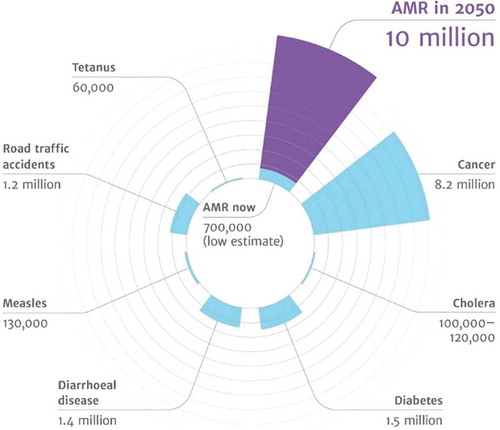
Figure 2. Overview of novel, nanotechnology-based antimicrobials, distinguishing organic and inorganic materials, together with their perceived advantages with respect to infection-control.
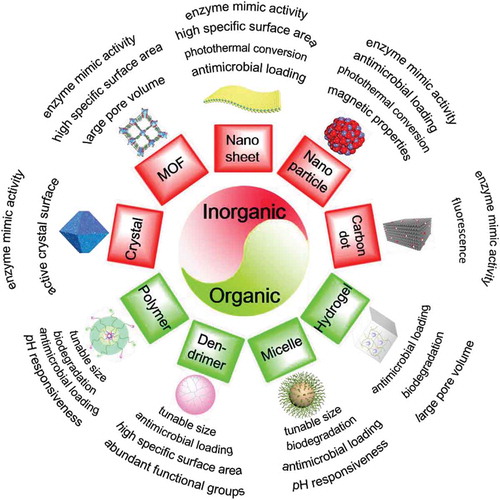
Figure 3. Examples of the advantages of nano-antimicrobials in combating infectious biofilms. (a) Fluorescence images showing the effect of a gold-nanorod coating on a glass surface in bacterial killing of an S. epidermidis biofilm by photothermal heat conversion. Biofilms on the gold-nanorod coated surface show almost complete bacterial killing (red-fluorescent bacteria) upon NIR-irradiation. Green-fluorescent bacteria are alive [Citation47] (with the permission of Elsevier Ltd.). (b) The idealized deep penetration and homogeneous distribution of magnetically targeted nanoparticles into infectious biofilms often assumed rather than experimentally demonstrated and not trivial to achieve [Citation49] (with permission of ACS Publications). (c) Penetration and accumulation of pH-responsive, Nile-red-loaded mixed-shell polymeric micelles in a staphylococcal biofilm at pH (5.0) [Citation64] (with permission of ACS Publications). (d) ROS production by graphene carbon dots (GQDs) upon sunlight irradiation (quantified by DCFH-DA staining) and E. coli survival (inset) [Citation30] (with permission from Taylor & Francis).
![Figure 3. Examples of the advantages of nano-antimicrobials in combating infectious biofilms. (a) Fluorescence images showing the effect of a gold-nanorod coating on a glass surface in bacterial killing of an S. epidermidis biofilm by photothermal heat conversion. Biofilms on the gold-nanorod coated surface show almost complete bacterial killing (red-fluorescent bacteria) upon NIR-irradiation. Green-fluorescent bacteria are alive [Citation47] (with the permission of Elsevier Ltd.). (b) The idealized deep penetration and homogeneous distribution of magnetically targeted nanoparticles into infectious biofilms often assumed rather than experimentally demonstrated and not trivial to achieve [Citation49] (with permission of ACS Publications). (c) Penetration and accumulation of pH-responsive, Nile-red-loaded mixed-shell polymeric micelles in a staphylococcal biofilm at pH (5.0) [Citation64] (with permission of ACS Publications). (d) ROS production by graphene carbon dots (GQDs) upon sunlight irradiation (quantified by DCFH-DA staining) and E. coli survival (inset) [Citation30] (with permission from Taylor & Francis).](/cms/asset/768bf56b-99d4-4279-9b0a-3435a7387911/iedd_a_1779697_f0003_oc.jpg)
Figure 4. Mechanisms of antibiotic-resistance and antibiotic targets. (a) Mechanisms of antimicrobial resistance, including biofilm formation and intrinsic mechanisms of antibiotic-resistance-based or natural resistance or resistance acquired through gene mutation of transfer. (B) Bacterial targets of different antibiotics and resistance mechanisms applied by bacterial pathogens [Citation77] (with permission from BioMed Central).
![Figure 4. Mechanisms of antibiotic-resistance and antibiotic targets. (a) Mechanisms of antimicrobial resistance, including biofilm formation and intrinsic mechanisms of antibiotic-resistance-based or natural resistance or resistance acquired through gene mutation of transfer. (B) Bacterial targets of different antibiotics and resistance mechanisms applied by bacterial pathogens [Citation77] (with permission from BioMed Central).](/cms/asset/46d34503-2dcc-407f-9314-4d4fcb46f152/iedd_a_1779697_f0004_oc.jpg)
Figure 5. (a) In vitro inhibition by different synthetic anhydrotetracycline (aTC) analogues of tetracycline destructase degradation as a function of inhibitor concentration, measured using an optical absorbance kinetic assay and expressed as a velocity parameter [Citation95]. (with permission from Springer). (b) Normalized nsaS expression as a part of the nisin efflux system in S. aureus SH1000 in a planktonic state and adhering to a hydrophobic polyethylene surface [Citation111,Citation115].
![Figure 5. (a) In vitro inhibition by different synthetic anhydrotetracycline (aTC) analogues of tetracycline destructase degradation as a function of inhibitor concentration, measured using an optical absorbance kinetic assay and expressed as a velocity parameter [Citation95]. (with permission from Springer). (b) Normalized nsaS expression as a part of the nisin efflux system in S. aureus SH1000 in a planktonic state and adhering to a hydrophobic polyethylene surface [Citation111,Citation115].](/cms/asset/b45f8010-7a4a-4cc9-bd6e-47d792ad0dbb/iedd_a_1779697_f0005_oc.jpg)
Figure 6. The concepts of mutant selection window (MSW) and mutant prevention concentration (MPC), as introduced by [Citation151]. (a) CFUs retrieved from a bacterial culture after antimicrobial exposure as a function of antibiotic concentration (Green and red indicate bacteria prior to and after mutation, respectively). (b) Serum concentration of an antibiotic as a function of time after administration, with MIC and MPC, indicated [adapted from reference 150] (with permission from Oxford university press).
![Figure 6. The concepts of mutant selection window (MSW) and mutant prevention concentration (MPC), as introduced by [Citation151]. (a) CFUs retrieved from a bacterial culture after antimicrobial exposure as a function of antibiotic concentration (Green and red indicate bacteria prior to and after mutation, respectively). (b) Serum concentration of an antibiotic as a function of time after administration, with MIC and MPC, indicated [adapted from reference 150] (with permission from Oxford university press).](/cms/asset/5e80017d-693d-4c3e-9115-ca95015f3d1f/iedd_a_1779697_f0006_oc.jpg)
Figure 7. Development of resistance against antibiotics upon bacterial exposure to nano-antimicrobials as compared with exposure to existing antibiotics. (a). Development resistance toward S. aureus exposed to quaternary ammonium modified gold nanoclusters (QA AuNCs) as compared with the development of resistance against oxacillin [Citation157]. (with permission from Wiley-VCH Verlag GmbH & Co). (b). Development of resistance toward P. aeruginosa exposed to poly(ε-caprolactone)-b-antimicrobial peptide coated Ag nanoparticles (AgNPs@PCL-b-AMPs) as compared with the development of resistance against ceftriaxone [Citation158] (with permission from ACS Publications).
![Figure 7. Development of resistance against antibiotics upon bacterial exposure to nano-antimicrobials as compared with exposure to existing antibiotics. (a). Development resistance toward S. aureus exposed to quaternary ammonium modified gold nanoclusters (QA AuNCs) as compared with the development of resistance against oxacillin [Citation157]. (with permission from Wiley-VCH Verlag GmbH & Co). (b). Development of resistance toward P. aeruginosa exposed to poly(ε-caprolactone)-b-antimicrobial peptide coated Ag nanoparticles (AgNPs@PCL-b-AMPs) as compared with the development of resistance against ceftriaxone [Citation158] (with permission from ACS Publications).](/cms/asset/6735a7b9-43da-41c0-9659-c4a2dd366079/iedd_a_1779697_f0007_oc.jpg)
