Figures & data
Figure 1. The MC4R pathway [Citation1–3]. LEPR, leptin receptor; MC4R, melanocortin-4 receptor; MSH, melanocyte-stimulating hormone; PCSK1, proprotein convertase subtilisin/kexin type 1; POMC, proopiomelanocortin.
![Figure 1. The MC4R pathway [Citation1–3]. LEPR, leptin receptor; MC4R, melanocortin-4 receptor; MSH, melanocyte-stimulating hormone; PCSK1, proprotein convertase subtilisin/kexin type 1; POMC, proopiomelanocortin.](/cms/asset/f373c271-187d-4c70-88e3-be8a8e7617d0/iere_a_2179985_f0001_oc.jpg)
Figure 2. In vitro functional assays for (A) LEPR, (B) PCSK1, and (C, D) POMC variants. AMC, 7-amino-4-methylcoumarin; cAMP, cyclic adenosine monophosphate; HEK, human embryonic kidney; LEPR, leptin receptor; MC4R, melanocortin-4 receptor; MSH, melanocyte-stimulating hormone; PCSK1, proprotein convertase subtilisin/kexin type 1; POMC, proopiomelanocortin; pSTAT3, phosphorylated signal transducer and activator of transcription 3; SRE, serum response element.
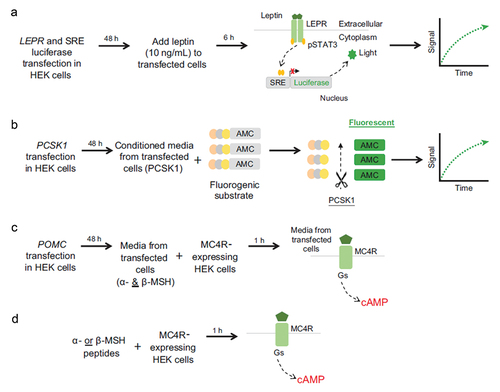
Table 1. Validation of assays against published variants of known functional impact.
Table 2. Criteria for variant classification.
Figure 3. Distributions of the mean percent of WT of all observed variants characterized in the functional assays. Each data point represents the mean (95% confidence interval in blue) of replicate assays and is compared with mean (95% confidence interval) results from replicate WT assays, shown in gray. Variants in red demonstrate a functional deficit, characterized by lack of overlap with confidence interval with results from WT assays. (A) All observed LEPR variants (n = 910). (B) All observed PCSK1 variants (n = 619). (C) All observed variants outside α-/β-MSH peptide region (n = 314). (D) Representation of variant by functional categorization for variants within the α-MSH peptide region (138–150; n = 17) and β-MSH region (213–234; n = 27). cAMP, cyclic adenosine monophosphate; MSH, melanocyte-stimulating hormone; pERTKR-AMC, Pyr-Glu-Arg-Thr-Lys-Arg-7-amino-4-methylcoumarin; STAT3, signal transducer and activator of transcription 3; WT, wild-type.
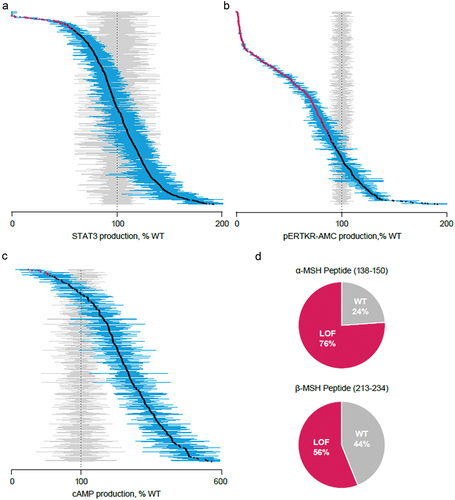
Table 3. List of observed variants for potential reclassification based on in vitro functional assays.
