Figures & data
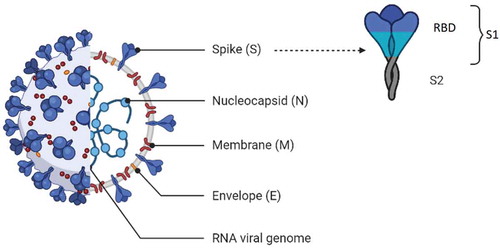
Figure 1. The main serological methods for detection of SARS‑CoV‑2 infection
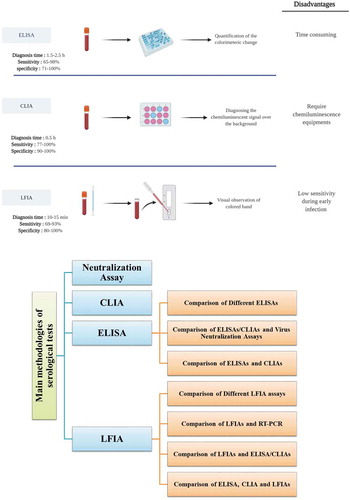
Figure 3. Mechanism of surrogate virus neutralization test (sVNT)
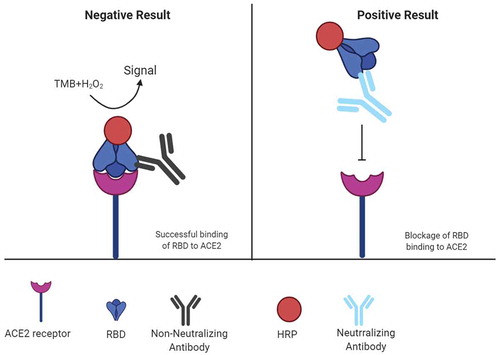
Figure 4. Electrochemiluminescence-based anti-SARS‑CoV‑2 immunoassay
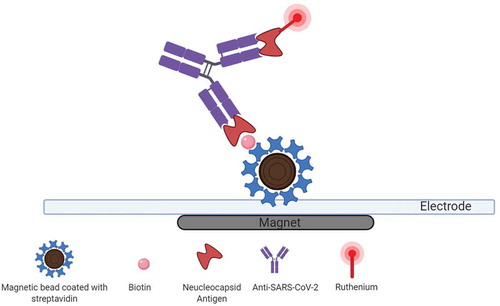
Table 1. Comparison of COVID-19 ELISA/CLIA and neutralization assays
Table 2. Comparison of COVID-19 ELISAs and CLIAs
Figure 5. Lateral flow immunoassays (LFIAs) for diagnosis of COVID-19
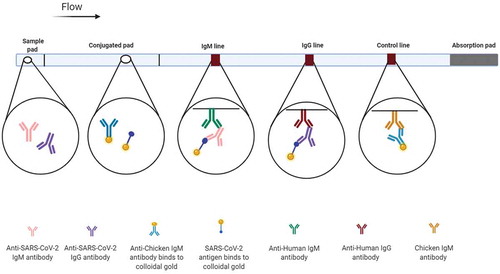
Table 3. Comparison of COVID-19 LFIAs and ELISA/CLIAs
Table 4. Comparison of COVID-19 ELISA, CLIA and LFIAs
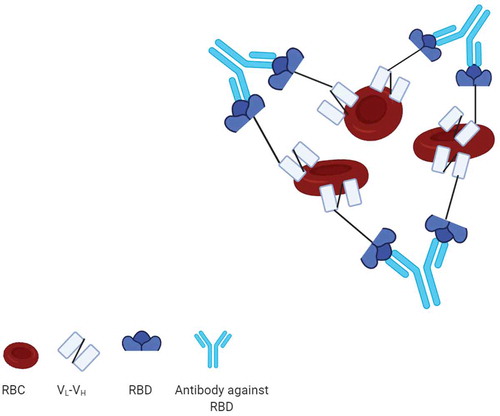
Figure 6. Mechanism of red blood cell agglutination assay