Figures & data
Table I. Summary of samples included in the survey. The catalogue numbers are vouchers deposited in the Bergen Museum, Natural History Collections of Bergen.
Table II. Taxon and authors for the species included in the analysis. GeneBank accession numbers for cytochrome c oxidase subunit I (COI) and the 16S sequences included in the combined analysis are also given.
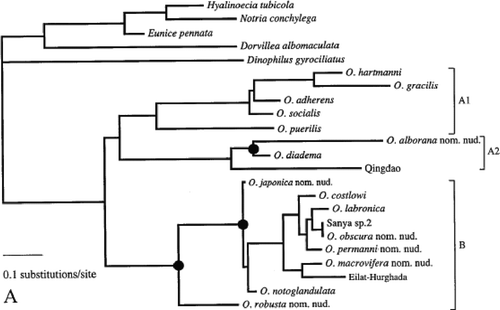
Figure 1. Consensus of the eight most-parsimonious trees resulting from the analysis of cytochrome c oxidase subunit I data.
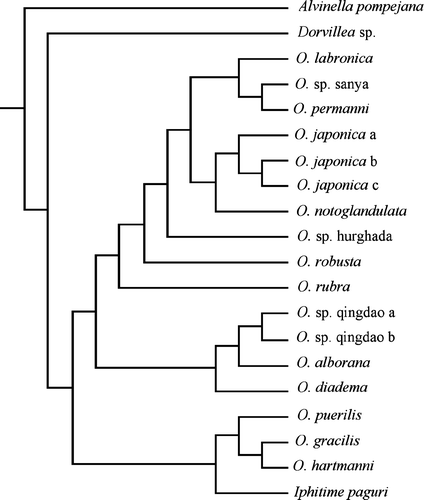
Figure 2. The single best tree from the maximum likelihood analysis of the cytochrome c oxidase subunit I data. Branch support values given in italics indicate bootstrap values, whereas those in bold are Bayesian values.
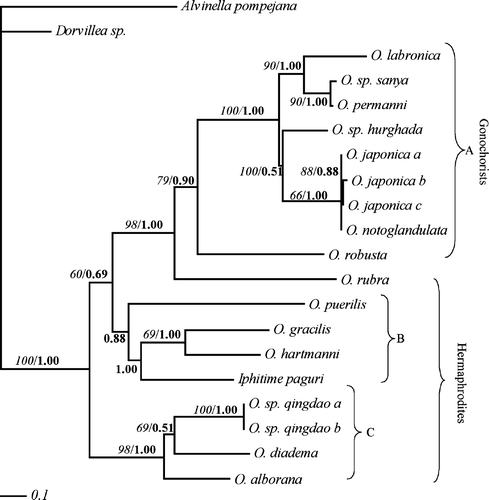
Figure 3. The single most-parsimonious tree resulting from the combined analysis including only the ophryotrochans.
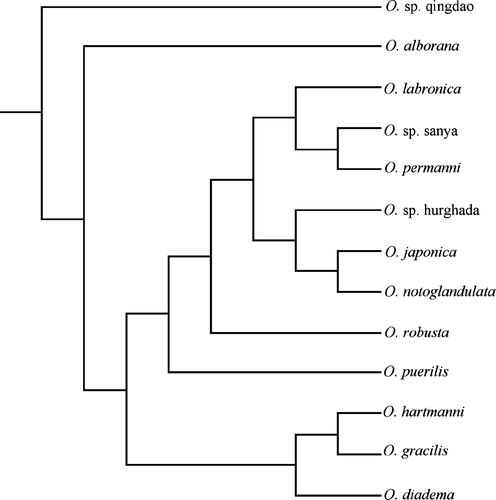
Figure 4. The single best likelihood tree from the combined analysis only including species from the genus Ophryotrocha. The branch values indicate bootstrap support. The line on the bottom left corner of the figure shows the number of substitutions.
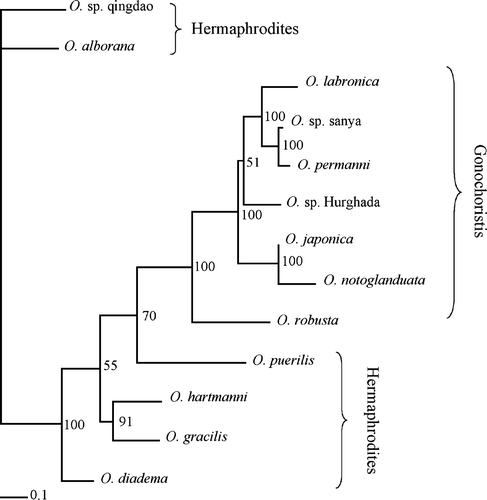
Figure 5. Bayesian consensus tree of the combined analysis. The values indicate posterior probabilities.
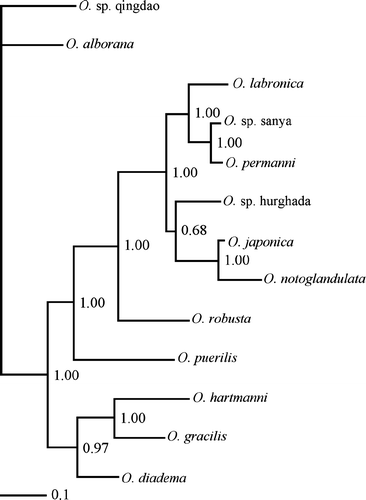
Figure 6. Phylogenetic tree resulting from the maximum likelihood analysis conducted by Dahlgren et al. (Citation2001). The filled circles indicate nodes congruent with the investigation of Pleijel & Eide (Citation1996).