Figures & data
Figure 1. Different length scales of cardiac tissue to be integrated into the scaffold: mm scale for vascularisation and nutrition, µm scale for cell accomodation, nm scale for the expression of ECM components.
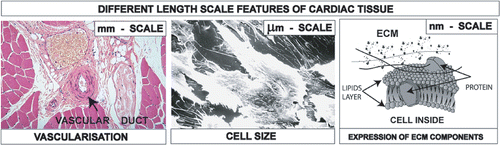
Figure 2. SEM micrographs of PLA scaffolds prepared by: (a) NaCl leaching technique, the inset reports a zoomed area of the scaffold; (b) paraffin leaching techniques, the inset reports the cross-sectional view.
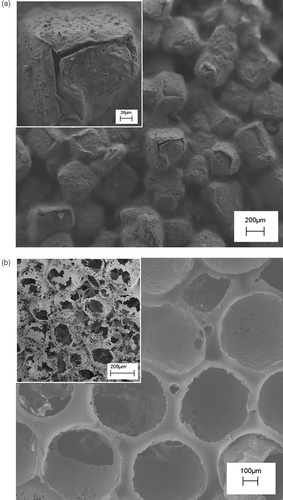
Figure 3. SEM images of a PLA scaffold prepared by the phase separation technique at two different temperature (T) of solvent extraction, (a) T = −18°C and (b) −18°C < T < 25°C. The insets show high magnification SEM micrographs of a PLA scaffold prepared by the phase separation technique at two different temperature (T) of solvent extraction, (a) T 1 = −18°C and (b) −18°C < T 2 < 25°C.
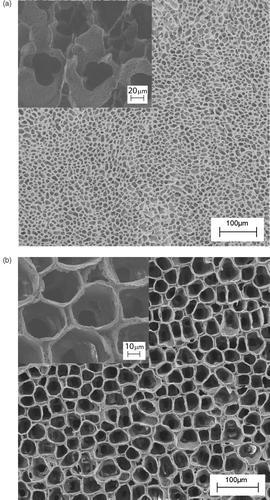
Figure 5. Schematics of the electrospinning experimental set up, where the grounded collecting electrodes were (a) two Cu parallel bars, (b) two pairs of Cu bar electrodes placed orthogonal to each other.
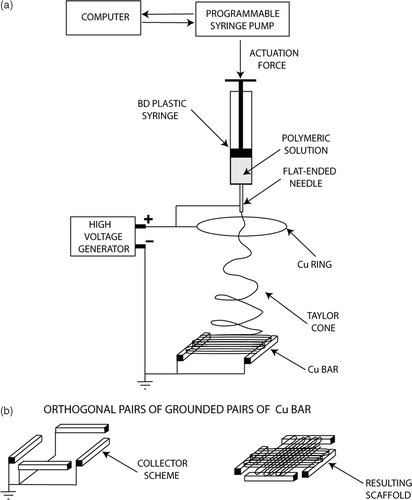
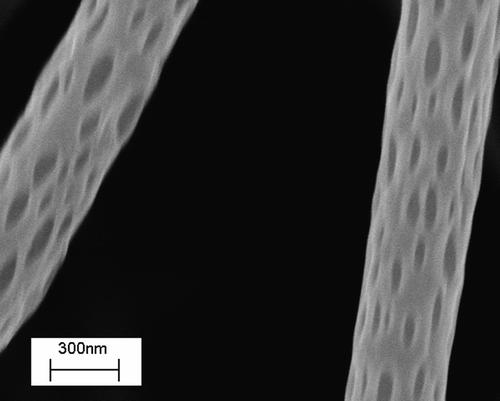
Figure 6. SEM micrographs of (a) oriented PCL fiber bundle, (b) macroscopically textured PCL scaffold.
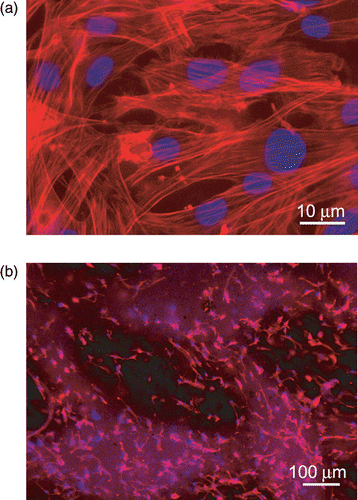
Figure 7. Immunofluorescence analysis of F-actin expression (red) in mMSC grown after 72 h, on (a) PLA scaffold shown in , (b) PCL scaffold shown in . Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Images of cell samples were taken with a Leica DMRB microscope using a digital camera. The images were representative of at least five random fields for each sample.