Figures & data
Figure 1. Interactivity for a protein-ligand system in the VR software Narupa IMD [Citation31,Citation32]. A) A protein-drug system (influenza neuraminidase complexed with the neuraminidase inhibitor oseltamivir) shown in the interactive molecular dynamics in virtual reality (IMD-VR) software Narupa IMD, where all atoms are rendered with the default ball and stick renderer. The VR controllers are also rendered in VR, where the orange circle represents the point of interaction with the controller (like a cursor for a mouse). B) Rendering of all atoms changed from ball and stick to lines and a different color scheme. C) The protein is here rendered in the ‘cartoon’ renderer and colored in rainbow, where red represents the N-terminal of the protein and violet represents the C-terminal. The drug is here rendered in the ‘cpm’ renderer and with atomistic colors to differentiate it from the protein. D) By grabbing single atoms of the drug molecule using each VR controller, the user can apply a force to coax the drug out of the active site. The force from the VR controllers is shown by the two light yellow sine waves between the controller and the drug molecule. E) The drug has now started to dissociate from the protein because of the interaction by the user in VR. F) A view of a VR user in real life, where the monitor is showing what the user sees in VR.
![Figure 1. Interactivity for a protein-ligand system in the VR software Narupa IMD [Citation31,Citation32]. A) A protein-drug system (influenza neuraminidase complexed with the neuraminidase inhibitor oseltamivir) shown in the interactive molecular dynamics in virtual reality (IMD-VR) software Narupa IMD, where all atoms are rendered with the default ball and stick renderer. The VR controllers are also rendered in VR, where the orange circle represents the point of interaction with the controller (like a cursor for a mouse). B) Rendering of all atoms changed from ball and stick to lines and a different color scheme. C) The protein is here rendered in the ‘cartoon’ renderer and colored in rainbow, where red represents the N-terminal of the protein and violet represents the C-terminal. The drug is here rendered in the ‘cpm’ renderer and with atomistic colors to differentiate it from the protein. D) By grabbing single atoms of the drug molecule using each VR controller, the user can apply a force to coax the drug out of the active site. The force from the VR controllers is shown by the two light yellow sine waves between the controller and the drug molecule. E) The drug has now started to dissociate from the protein because of the interaction by the user in VR. F) A view of a VR user in real life, where the monitor is showing what the user sees in VR.](/cms/asset/1618e02c-a5c8-484d-b8a3-2ff95c78460e/iedc_a_2079632_f0001_oc.jpg)
Figure 2. VR Gloves and their application [Citation54]. (A) Pinch sensing gloves. The tracker is placed on top of the glove and the interaction occurs through a pinching motion between either the thumb and index finger, or thumb and middle finger. (B) Two users in VR interacting with a real-time MD simulation of a polyalanine peptide. A pinching motion allows the users to grab an atom and interactively maneuver the protein around the VR environment. Image reproduced from [Citation54] licensed under CC BY-SA.
![Figure 2. VR Gloves and their application [Citation54]. (A) Pinch sensing gloves. The tracker is placed on top of the glove and the interaction occurs through a pinching motion between either the thumb and index finger, or thumb and middle finger. (B) Two users in VR interacting with a real-time MD simulation of a polyalanine peptide. A pinching motion allows the users to grab an atom and interactively maneuver the protein around the VR environment. Image reproduced from [Citation54] licensed under CC BY-SA.](/cms/asset/8b1bd4ec-d8e7-48c3-9583-e7ac13163fa2/iedc_a_2079632_f0002_oc.jpg)
Figure 3. Scenes from VR software for molecular modeling. 1) A scene from the interactive molecular dynamics tutorial from the Nanome software [Citation30], where the user is inspecting the first frame of a trajectory. 2) UnityMol software [Citation103] showing a cartoon and hyperball representation of the influenza neuraminidase protein (PDB 3TI6). 3) Hydrogen bonding (rendered as white glowing interactions) between a 20 amino acid long polypeptide in the software, Peppy [Citation88]. 4) The SARS-CoV Mpro (PDB 2Q6G) in a rainbow cartoon representation using the web-based VR software ProteinVR [Citation29]. Images created by the author using interactive VR software Nanome [Citation30], UnityMol [Citation103], Peppy [Citation88], and ProteinVR [Citation29].
![Figure 3. Scenes from VR software for molecular modeling. 1) A scene from the interactive molecular dynamics tutorial from the Nanome software [Citation30], where the user is inspecting the first frame of a trajectory. 2) UnityMol software [Citation103] showing a cartoon and hyperball representation of the influenza neuraminidase protein (PDB 3TI6). 3) Hydrogen bonding (rendered as white glowing interactions) between a 20 amino acid long polypeptide in the software, Peppy [Citation88]. 4) The SARS-CoV Mpro (PDB 2Q6G) in a rainbow cartoon representation using the web-based VR software ProteinVR [Citation29]. Images created by the author using interactive VR software Nanome [Citation30], UnityMol [Citation103], Peppy [Citation88], and ProteinVR [Citation29].](/cms/asset/82351b45-c7af-4679-8616-d559fd3b4435/iedc_a_2079632_f0003_oc.jpg)
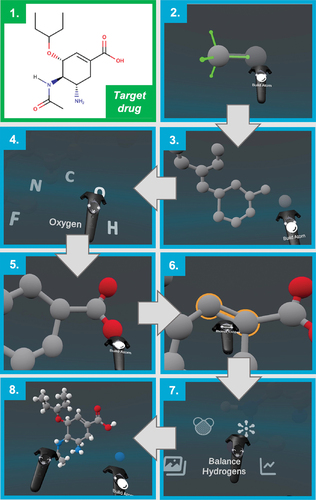
Figure 4. Constructing oseltamivir using the Narupa Builder. 1) The structure of the target drug (oseltamivir carboxylate, commonly known as Tamiflu, an influenza drug) to be built using Narupa Builder. 2) Building the first carbon atoms of the 6 membered ring. The green axes represent the possible sites for a new atom to be bonded to the carbon. 3) The carbon backbone of the drug, without any hydrogens. 4) Changing from a carbon atom to an oxygen atom to be added to the drug. 5) Creating the carboxylate functional group. 6) Changing the bond order between two carbon atoms from a single to a double bond. 7) Finally, adding hydrogens to all the possible sites of hydrogenation. 8) The resulting structure.