Figures & data
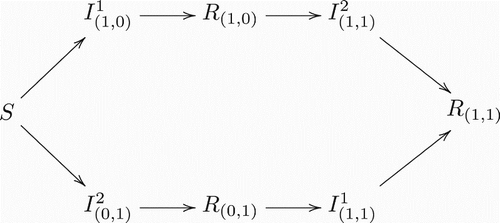
Figure 1. Transitions between classes when there is only one strain present. Notice this is a regular Susceptible-Infected-Recovered model.
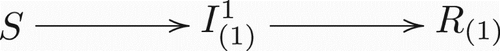
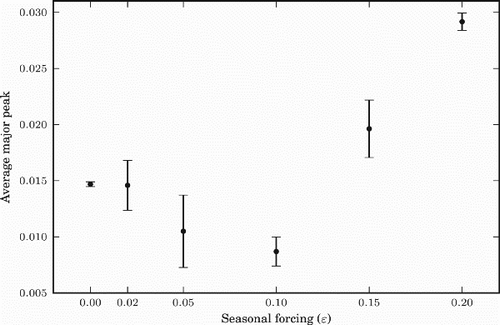
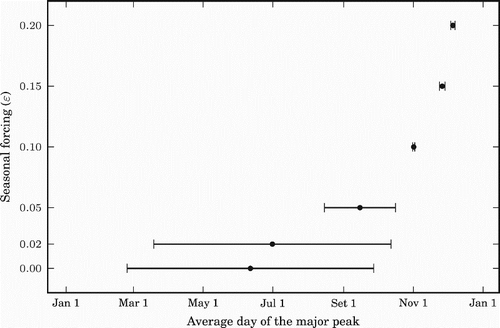
Figure 6. Prevalence (measured in percentage of the population infected per day) for different values of non-negative ϵ, i.e. for the tropics and the northern hemisphere. The dashed vertical lines represent the middle of the winter of each year.
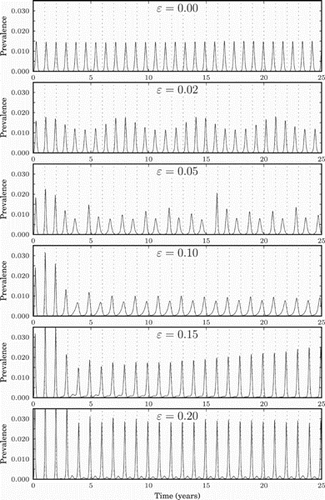
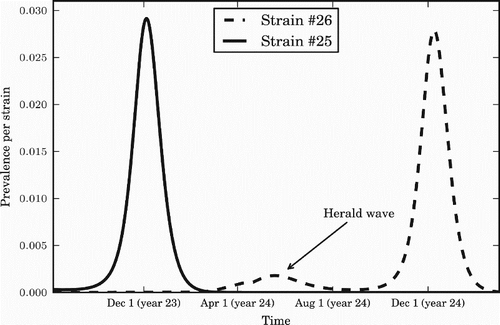
Figure 7. The herald wave phenomenon. A summer wave of a new strain predicts the strain that will dominate over the following winter. The value of seasonal forcing used is ϵ=0.20. Prevalence for each strain is measured in percentage of the population infected per day.