Figures & data
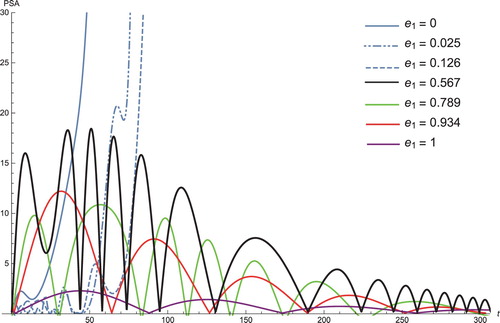
Figure 1. The solution profiles of , the androgen-dependent cancer cells. 1: MDDIM, 2: QSS, Quasi-steady state approximation, 3: numerical simulations for the full model. These cancer cells grow at the beginning of the treatment, then after they decrease very quickly.
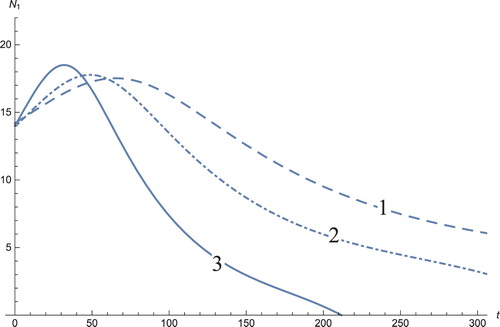
Figure 2. The solution profiles of , the androgen-independent cancer cells. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model. These cells decrease rapidly with the onset of immunotherapy.
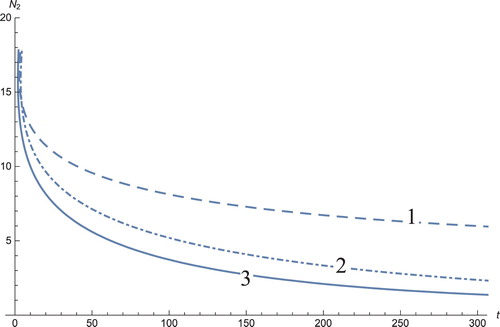
Figure 3. The solutions profiles of the T cells. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model. At first treatment, there is a very sharp increase in these cells and then drop to equilibrium point.
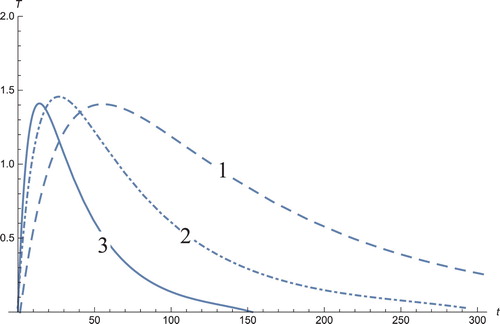
Figure 4. The solution profiles of , the concentration of cytokine. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model. During the treatment, the concentration of cytokines increases due to the intense activity of the immune system.
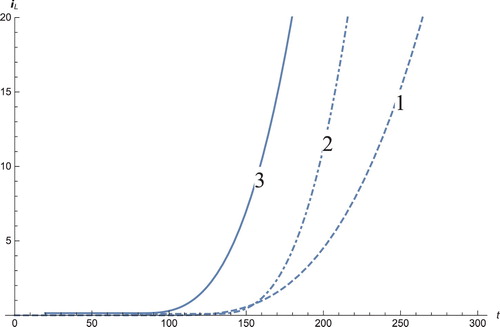
Figure 5. The solution profiles of , the concentration of androgen. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model. The androgens corresponding to the dendritic cells stabilize after ≈60 days, to their equilibrium point.
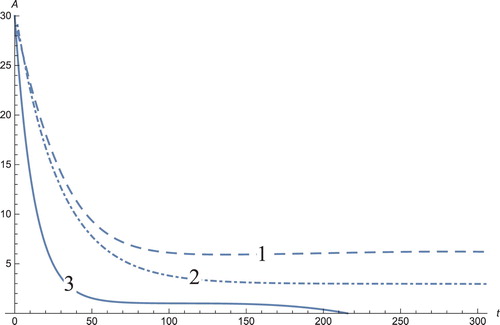
Figure 6. The solution profiles of D, the number of dendritic cells. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model. The number of dendritic cells stabilizes after days.
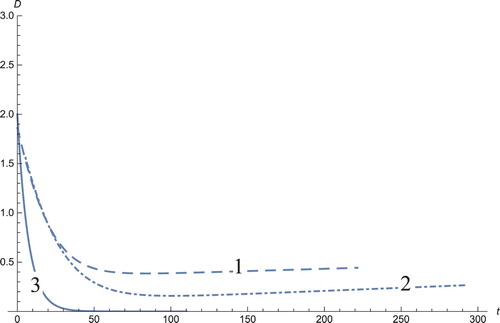
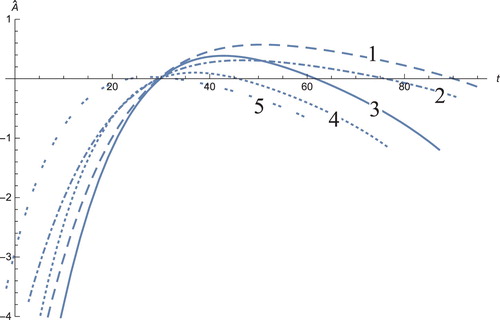
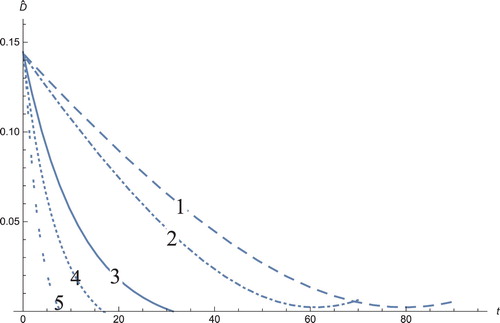
Figure 7. The solution profiles of , the new variable in the new coordinates. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model in the new coordinates, 4: combination of MDDIM and SPVF, 5: SPVF method.
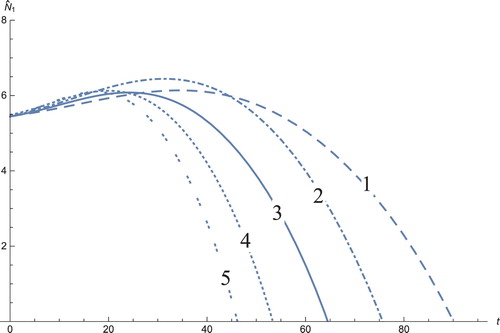
Figure 8. The solution profiles of , the new variable in the new coordinates. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model in the new coordinates, 4: combination of MDDIM and SPVF, 5: SPVF method.
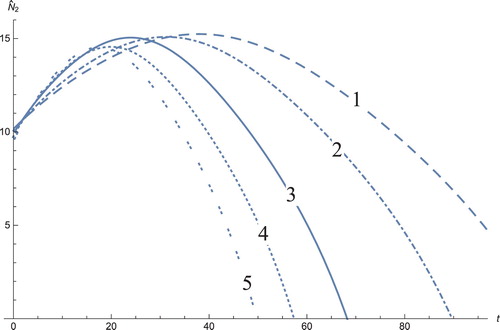
Figure 9. The solution profiles of , the new variable in the new coordinates. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model in the new coordinates, 4: combination of MDDIM and SPVF, 5: SPVF method.
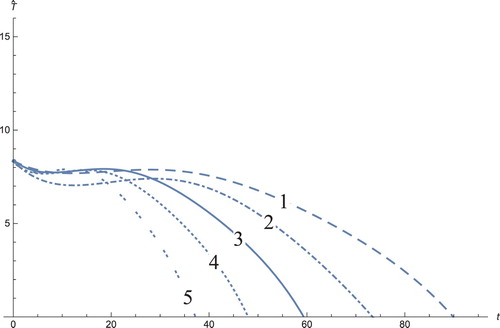
Figure 10. The solution profiles of , the new variable in the new coordinates. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model in the new coordinates, 4: combination of MDDIM and SPVF, 5: SPVF method.
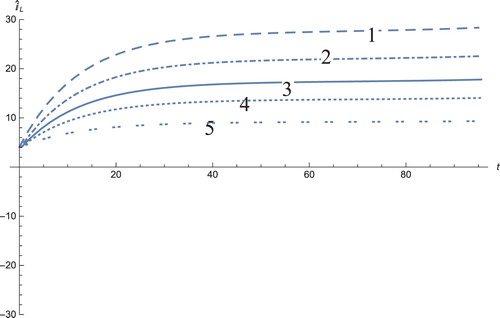
Figure 11. The solution profiles of , the new variable in the new coordinates. 1: MDDIM, 2: QSS, quasi-steady state approximation, 3: numerical simulations for the full model in the new coordinates, 4: combination of MDDIM and SPVF, 5: SPVF method. Since we transfer the model using eigenvectors, we obtain a negative value of
which is biologically insignificant. And hence, for sake of biological understanding, we must ignore these negative values.