Figures & data
Figure 1. The uniqueness of the final epidemic size, with respect to the basic reproduction number .
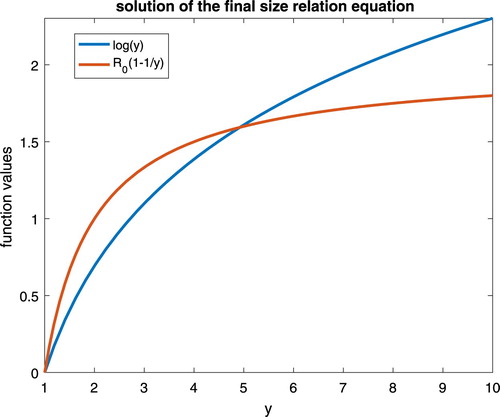
Figure 2. The x-axis represents time (in days) and the y-axis represents the numbers of individuals in different compartments with respect to time. Outbreaks are initiated by one infectious individual. Plots on lower left and on lower right represent the sums of ,
and
for models (Equation12
(12)
(12) ) and (Equation19
(19)
(19) ), respectively.
![Figure 2. The x-axis represents time (in days) and the y-axis represents the numbers of individuals in different compartments with respect to time. Outbreaks are initiated by one infectious individual. Plots on lower left and on lower right represent the sums of S1+S2, I1+I2 and R1+R2 for models (Equation12(12) S1′=−S1[p11β1I1N1+p12β1I2N2],I1′=S1[p11β1I1N1+p12β1I2N2]−γI1,R1′=γI1,S2′=−S2[p21β2I1N1+p22β2I2N2],I2′=S2[p21β2I1N1+p22β2I2N2]−γI2,R2′=γI2.(12) ) and (Equation19(19) S1′=−S1a1(t)[p1I1N1+p2I2N2],I1′=S1a1(t)[p1I1N1+p2I2N2]−γI1,R1′=γI1,S2′=−S2a2(t)[p1I1N1+p2I2N2],I2′=S2a2(t)[p1I1N1+p2I2N2]−γI2,R2′=γI2.(19) ), respectively.](/cms/asset/c9268d44-6c21-4283-9644-e82cd4e1ac54/tjbd_a_1853833_f0002_oc.jpg)
Table 1. The final epidemic sizes for epidemic models (Equation3(3)
(3) ), (Equation7
(7)
(7) ), (Equation12
(12)
(12) ) and (Equation19
(19)
(19) ).
Figure 3. The comparison of final sizes predicted by the epidemic model (Equation3(3)
(3) ) with homogeneous mixing and the epidemic models (Equation12
(12)
(12) ) with heterogeneous mixing, with different combinations of parameters. All models share the identical basic reproduction number
.
![Figure 3. The comparison of final sizes predicted by the epidemic model (Equation3(3) S′=−βSIN,I′=βSIN−γI,R′=γI.(3) ) with homogeneous mixing and the epidemic models (Equation12(12) S1′=−S1[p11β1I1N1+p12β1I2N2],I1′=S1[p11β1I1N1+p12β1I2N2]−γI1,R1′=γI1,S2′=−S2[p21β2I1N1+p22β2I2N2],I2′=S2[p21β2I1N1+p22β2I2N2]−γI2,R2′=γI2.(12) ) with heterogeneous mixing, with different combinations of parameters. All models share the identical basic reproduction number R0.](/cms/asset/09a893dd-6a81-4cb6-9499-75618184ed96/tjbd_a_1853833_f0003_oc.jpg)
Figure 4. Five realizations generated by SSA for CTMC model (Equation21(21)
(21) ) with homogeneous mixing.
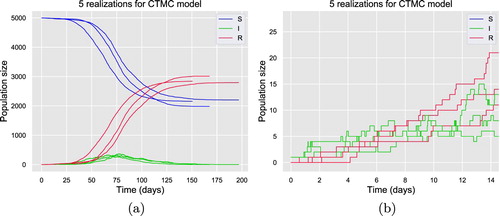
Figure 5. Five realizations generated by SSA for CTMC model (Equation24(24)
(24) ) with heterogeneous mixing. The top two subfigures are with initial condition of 1 infectious superspreader. The bottom two subfigures are with initial condition of 1 infectious non-superspreader.
![Figure 5. Five realizations generated by SSA for CTMC model (Equation24(24) P((S1,t+Δt,I1,t+Δ)−(S1,t,I1,t)=(−1,1))=S1,tβ1[p1I1,tN1+p2I2,tN2]+o(Δ),P((I1,t+Δt,R1,t+Δ)−(I1,t,R1,t)=(−1,1))=γI1,t+o(δ),P((S2,t+Δt,I2,t+Δ)−(S2,t,I2,t)=(−1,1))=S2,tβ2[p1I1,tN1+p2I2,tN2]+o(Δ),P((I2,t+Δt,R2,t+Δ)−(I2,t,R2,t)=(−1,1))=γI2,t+o(Δ)(24) ) with heterogeneous mixing. The top two subfigures are with initial condition of 1 infectious superspreader. The bottom two subfigures are with initial condition of 1 infectious non-superspreader.](/cms/asset/88dc090b-091e-4db7-afc1-20732450f7f2/tjbd_a_1853833_f0005_oc.jpg)
Figure 6. The distribution of final epidemic sizes for realizations for CTMC model (Equation21
(21)
(21) ) with homogeneous mixing.
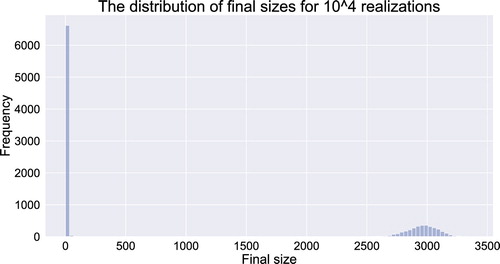
Figure 7. The distributions of final epidemic sizes for realizations for CTMC model (Equation24
(24)
(24) ) with heterogeneous mixing,
initiated by 1 infectious superspreader and
initiated by 1 infectious non-superspreader.
![Figure 7. The distributions of final epidemic sizes for 104 realizations for CTMC model (Equation24(24) P((S1,t+Δt,I1,t+Δ)−(S1,t,I1,t)=(−1,1))=S1,tβ1[p1I1,tN1+p2I2,tN2]+o(Δ),P((I1,t+Δt,R1,t+Δ)−(I1,t,R1,t)=(−1,1))=γI1,t+o(δ),P((S2,t+Δt,I2,t+Δ)−(S2,t,I2,t)=(−1,1))=S2,tβ2[p1I1,tN1+p2I2,tN2]+o(Δ),P((I2,t+Δt,R2,t+Δ)−(I2,t,R2,t)=(−1,1))=γI2,t+o(Δ)(24) ) with heterogeneous mixing, (a) initiated by 1 infectious superspreader and (b) initiated by 1 infectious non-superspreader.](/cms/asset/c94c5354-8bef-4533-a5f9-56a194f9a844/tjbd_a_1853833_f0007_oc.jpg)
Table 2. The probability of a minor outbreak for CTMC models (Equation21(21)
(21) ) with initially 1 infectious individual, and (Equation24
(24)
(24) ) with initially 1 infectious super-spreader and 1 infectious non super-spreader, respectively. It is demonstrated that, even with identical
, the probability of a minor outbreak can be varied by different choice of epidemic models.
