Figures & data
Figure 1. Diagram for an SEIR-type model with an Erlang latent period and Coxian infectious period. A special case of Equations (Equation15(15d)
(15d) ) and Equations (Equation20
(20d)
(20d) ). See the main text for details (and compare to Figure in [Citation83]).
![Figure 1. Diagram for an SEIR-type model with an Erlang latent period and Coxian infectious period. A special case of Equations (Equation15(15d) dRdt=(−AI1)Ty⏞(15d) ) and Equations (Equation20(20d) dRdt=rIIkI+∑j=1kI−1(1−ρIj)λIjIj.(20d) ). See the main text for details (and compare to Figure 2 in [Citation83]).](/cms/asset/473ca429-da57-4c8f-82f5-e759f33c06ad/tjbd_a_1912418_f0001_ob.jpg)
Figure 2. SEIR-type model with heterogeneity in illness severity and hospitalizations that do not alter the infectious period. A special case of Equations (Equation21(21e)
(21e) ) (cf. Figure ). See the main text for details. Here the standard LCT has been applied to the exposed (E) state, and the GLCT is applied to the I states, using the competing Poisson process approach [Citation51] to model hospitalizations in a fraction of the I state individuals. This ensures that the time spent in I is independent of whether or not individuals transition to the hospital or not. Compare to [Citation35].
![Figure 2. SEIR-type model with heterogeneity in illness severity and hospitalizations that do not alter the infectious period. A special case of Equations (Equation21(21e) dRdt=(−AI01)Ty0+(−AIs1)Tys.(21e) ) (cf. Figure 3). See the main text for details. Here the standard LCT has been applied to the exposed (E) state, and the GLCT is applied to the I states, using the competing Poisson process approach [Citation51] to model hospitalizations in a fraction of the I state individuals. This ensures that the time spent in I is independent of whether or not individuals transition to the hospital or not. Compare to [Citation35].](/cms/asset/36cb12cb-feca-4db5-98db-5ad51def7d2d/tjbd_a_1912418_f0002_ob.jpg)
Figure 3. SEIR-type model with heterogeneity in illness severity in which a fraction of infected individuals experience severe illness, and a fraction of those require critical care. This is also a special case of Equations (Equation21(21e)
(21e) ) (see Figure ). See the main text for details.
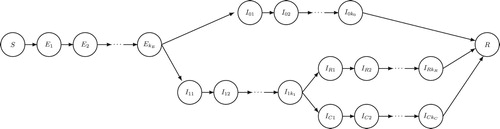
Figure 4. Benchmark results for 140 iterations computing numerical solutions to Rosenzweig-MacArthur model with Erlang (gamma) distributed maturation times and time spent in the adult-stage, using either a direct (LCT; medium grey) or more general (GLCT; black) formulations of the model (the standard Rosenzweig-MacArthur model with no maturation delay and exponentially distributed time spent in the adult stage [light grey; Equations (Equation8(8b)
(8b) )] is also included as a baseline). The second and third cases are mathematically equivalent systems. For smaller shape parameters (lower dimension systems) the GLCT model formulation is relatively slower than explicitly writing out the 2M + 1 equations, whereas for larger shape parameters (higher dimension systems) the GLCT-based formulation becomes markedly faster. This is likely due to the efficiency of the matrix computations. The x-axis values M indicate the number of maturing predator sub-states (
) and adult predator sub-states (
), which yields a 2M + 1 dimensional model. Numerical solutions were computed using the ode() function in the deSolve package [Citation101] in R [Citation90], using method ode45 with atol=10-6, for time points 0 to 500 in increments of 1, and model parameters r = 1, K = 1000, a = 5, h = 500,
,
,
, with initial conditions
,
(
),
, and
(j>1).
![Figure 4. Benchmark results for 140 iterations computing numerical solutions to Rosenzweig-MacArthur model with Erlang (gamma) distributed maturation times and time spent in the adult-stage, using either a direct (LCT; medium grey) or more general (GLCT; black) formulations of the model (the standard Rosenzweig-MacArthur model with no maturation delay and exponentially distributed time spent in the adult stage [light grey; Equations (Equation8(8b) dPdt=χaNh+NP−μP(8b) )] is also included as a baseline). The second and third cases are mathematically equivalent systems. For smaller shape parameters (lower dimension systems) the GLCT model formulation is relatively slower than explicitly writing out the 2M + 1 equations, whereas for larger shape parameters (higher dimension systems) the GLCT-based formulation becomes markedly faster. This is likely due to the efficiency of the matrix computations. The x-axis values M indicate the number of maturing predator sub-states (kx=M) and adult predator sub-states (ky=M), which yields a 2M + 1 dimensional model. Numerical solutions were computed using the ode() function in the deSolve package [Citation101] in R [Citation90], using method ode45 with atol=10-6, for time points 0 to 500 in increments of 1, and model parameters r = 1, K = 1000, a = 5, h = 500, χ=0.5, μx=0.5, μy=1, with initial conditions N(0)=1000, xi(0)=0 (i≥1), y1(0)=10, and yj(0)=0 (j>1).](/cms/asset/4e72f71e-3278-4a76-a7a2-147a7b514578/tjbd_a_1912418_f0004_ob.jpg)
Figure 5. Benchmark results for 500 iterations computing numerical solutions to an SEIR model with latent and infectious period distributions that are either Erlang (panel a; Equation (Equation18(18d)
(18d) )) or Coxian (panel b; Equation (Equation20
(20d)
(20d) )), using either a direct equation-by-equation formulation (LCT; medium grey) or more general matrix-vector formulation (GLCT; black) of the model (SEIR with exponentially distributed latent and infectious periods [light grey; Equation (14)] also included as a baseline). The second and third cases are mathematically equivalent systems. For smaller shape parameters (lower dimension systems) the GLCT model formulation is relatively slower than explicitly writing out the 2N + 2 equations, whereas for larger shape parameters (higher dimension systems) the GLCT-based formulation becomes faster, presumably due to the efficiency of the matrix computations. The x-axis values N indicate the number of sub-states in each of E (
) and I (
), which yields a 2N + 2 dimensional model. Numerical solutions were computed using the ode() function in the deSolve package [Citation101] in R [Citation90], using method ode45 with atol=10-6, for time points 0 to 100 in increments of 0.5, and model parameters
,
,
, where all
, with initial conditions
,
,
(i>1),
,
(i>1), and
. See Supplementary Materials for more details.
![Figure 5. Benchmark results for 500 iterations computing numerical solutions to an SEIR model with latent and infectious period distributions that are either Erlang (panel a; Equation (Equation18(18d) dRdt=kIτIIkI(18d) )) or Coxian (panel b; Equation (Equation20(20d) dRdt=rIIkI+∑j=1kI−1(1−ρIj)λIjIj.(20d) )), using either a direct equation-by-equation formulation (LCT; medium grey) or more general matrix-vector formulation (GLCT; black) of the model (SEIR with exponentially distributed latent and infectious periods [light grey; Equation (14)] also included as a baseline). The second and third cases are mathematically equivalent systems. For smaller shape parameters (lower dimension systems) the GLCT model formulation is relatively slower than explicitly writing out the 2N + 2 equations, whereas for larger shape parameters (higher dimension systems) the GLCT-based formulation becomes faster, presumably due to the efficiency of the matrix computations. The x-axis values N indicate the number of sub-states in each of E (kE=N) and I (kI=N), which yields a 2N + 2 dimensional model. Numerical solutions were computed using the ode() function in the deSolve package [Citation101] in R [Citation90], using method ode45 with atol=10-6, for time points 0 to 100 in increments of 0.5, and model parameters β=1, μE=4, μI=7, where all ρE=ρI=0.99, with initial conditions S(0)=0.9999, E1(0)=0.0001, Ei(0)=0 (i>1), I1(0)=0, Ii(0)=0 (i>1), and R(0)=0. See Supplementary Materials for more details.](/cms/asset/19b61798-ac82-4f47-b3a1-1182a64ca2c1/tjbd_a_1912418_f0005_ob.jpg)
