Figures & data
Table 1. Descriptions of variables used in the model system (Equation1(1)
(1) ).
Figure 1. Schematic diagram of model system (Equation1(1)
(1) ).
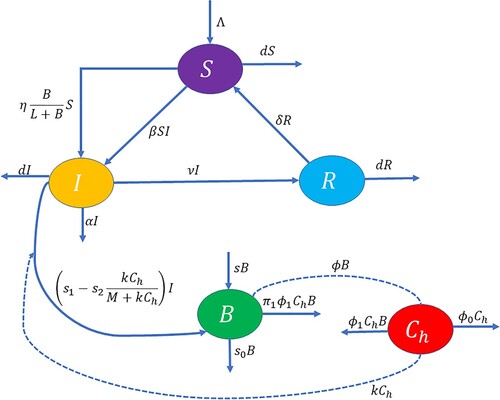
Table 2. Descriptions of parameters involved in system (Equation1(1)
(1) ).
Table 3. Values of parameters used for numerical simulations of system (Equation1(1)
(1) ).
Figure 2. The normalized forward sensitivity indices of the basic reproduction number () with respect to the model parameters Λ, β, η, L, ν,
,
and s. Here, the values of parameters are chosen from Table .
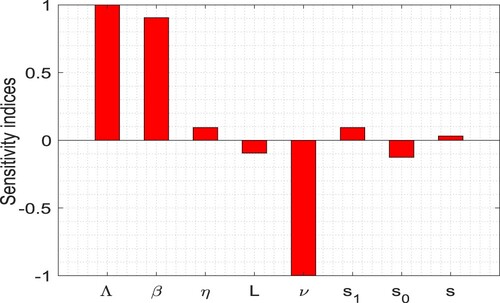
Figure 3. The uncertainty of the model (Equation1(1)
(1) ) on infected individuals (I). Baseline values of parameters are the same as in Table except
. Significant parameters are marked by
(p-value
).
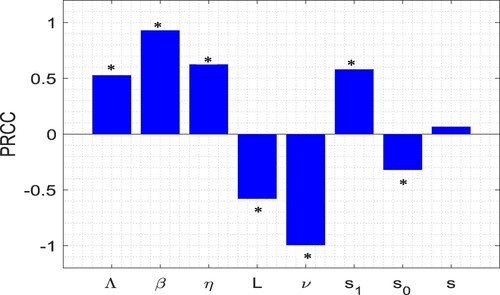
Figure 4. Contour plots of the basic reproduction number () as a functions of
and ν. Rest of the parameters are at the same values as in Table . In the figure, dashed green line stands for
. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
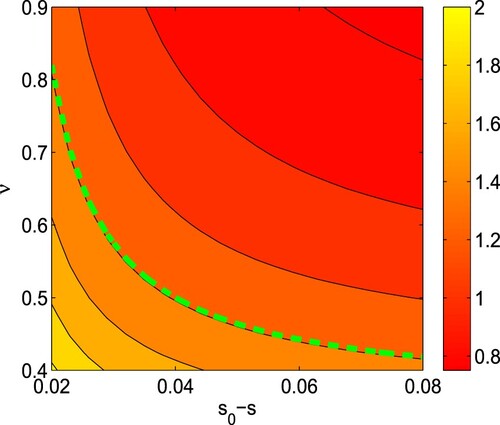
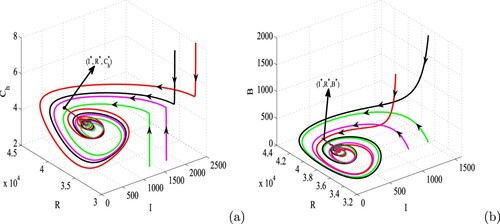
Figure 5. Bifurcation diagram of system (Equation1(1)
(1) ) with respect to
. Parameters are at the same values as in Table except
, d = 0.004,
and
. Here, blue, red and magenta colours, respectively, denote the stable disease-free equilibrium (
), unstable disease-free equilibrium (
) and the stable endemic equilibrium (
). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
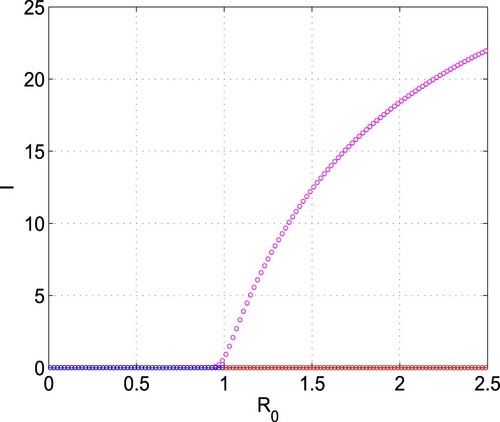
Figure 6. Surface plots of the infective population (first column) and bacterial density (second column) with respect to (a) β and , and (b) η and
. Other parameters are at the same values as in Table .
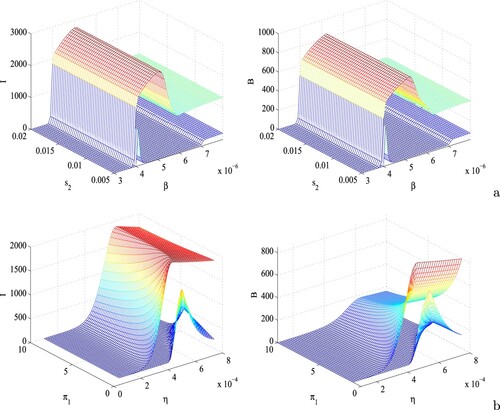
Figure 7. Surface plots of the infective population (first column) and bacterial density (second column) with respect to (a) ϕ and , and (b)
and k. Other parameters are at the same values as in Table .
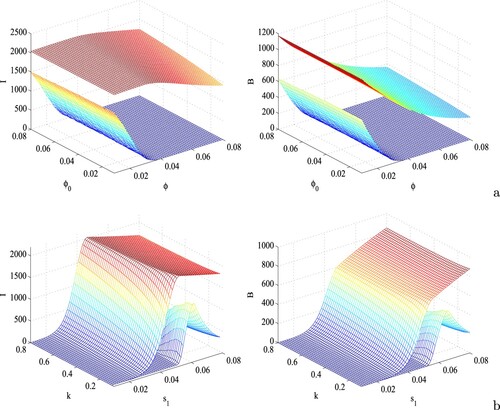
Figure 8. Variations in the infective population (first column) and bacterial density (second column) with respect to time for different combinations of (a) and
, and (b) k and
. Rest of the parameters are at the same values as in Table except
.
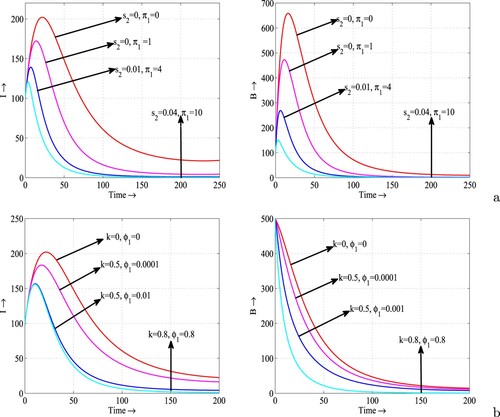
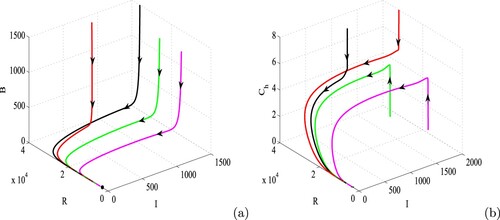
Figure 9. Figure illustrating the global asymptotic stability of disease-free equilibrium in (a) I−R−B and (b)
spaces. Parameters are at the same values as in Table except
, d = 0.004,
and
. Rest of the parameters are at the same values as in Table .