Figures & data
Figure 1. Location of the Seurasaari Island for test and snapshots of (a) the WPI covering the island and (b) the TLS-collected point clouds at the centre of the island.
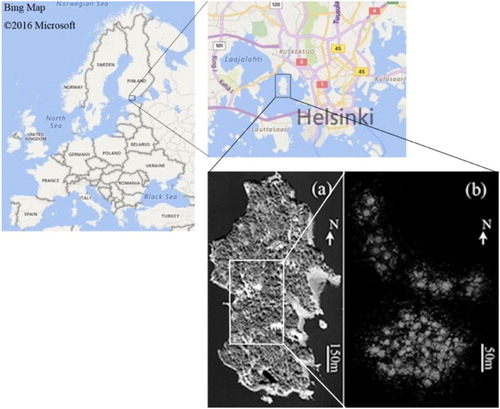
Table 1. Statistical characteristics of the individual sample trees selected for the test.
Figure 2. Schematic diagram of defining the proposed SSVs: DBH, FLBH, SS and TH (the two dotted lines are parallel).
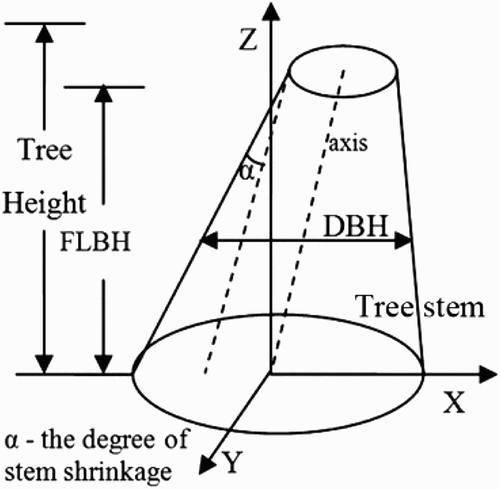
Table 2. Definitions of the proposed texture-kind feature parameters based on WPIs.
Figure 3. Scatterplots of the TLS-derived and manually measured DBH values for the 62 sample trees for the test.
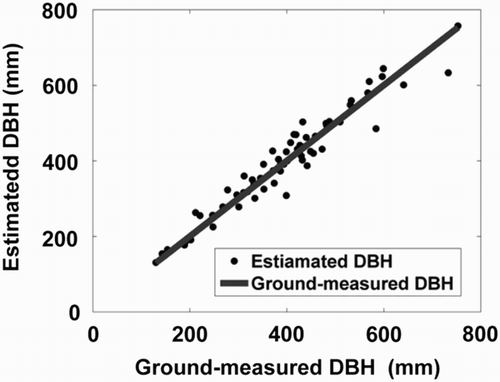
Figure 4. Boxplots of the WPI-derived feature parameters: (a) area of crown, (b) ED and (c) the ratio between the area of crown and the area of MBR.
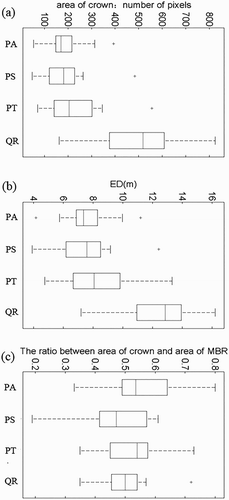
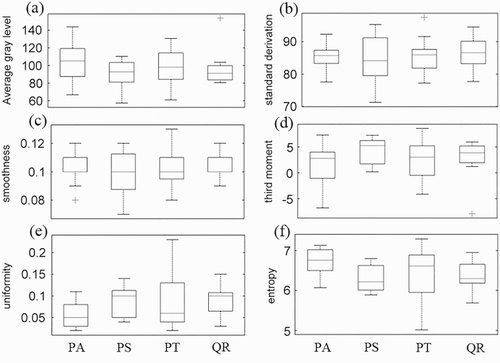
Figure 5. Boxplots of the WPI-derived SDGL-typed feature parameters: (a) AV, (b) SD, (c) SM, (d) TM, (e) UN and (f) ET.