Figures & data
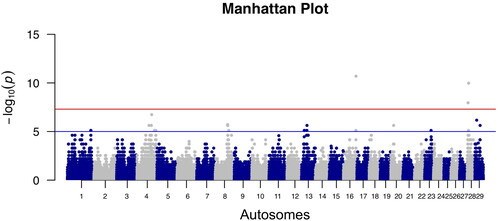
Table 1. Genome Wide Association Study (GWAS) results: SNPs over the Bonferroni (bold) and False Discovery Rate (FDR) genome wide thresholds (0.05 significance value) together with candidate genes.
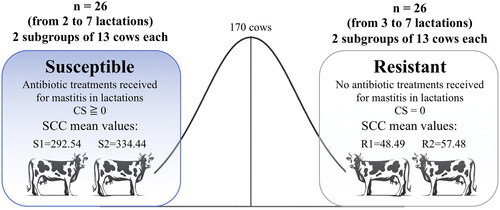
Figure 1. Experimental design scheme: Susceptible (S1 and S2) and Resistant (R1 and R2) cows in a Selective genotyping and pooling approach.
Supplemental Material
Download MS Word (17.9 KB)Supplemental Material
Download MS Excel (23.4 KB)Supplemental Material
Download MS Word (22.1 KB)Data availability statement
The data are still under embargo as part of a project still active.