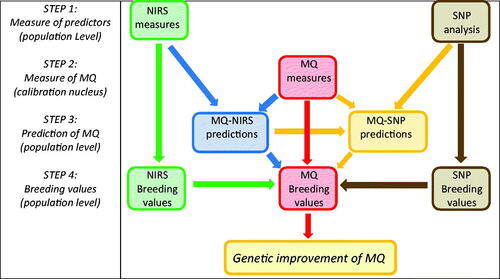
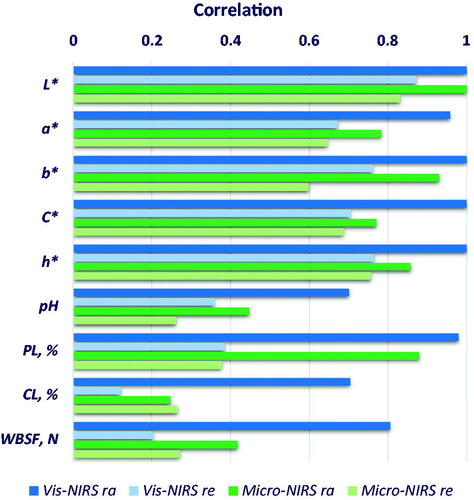
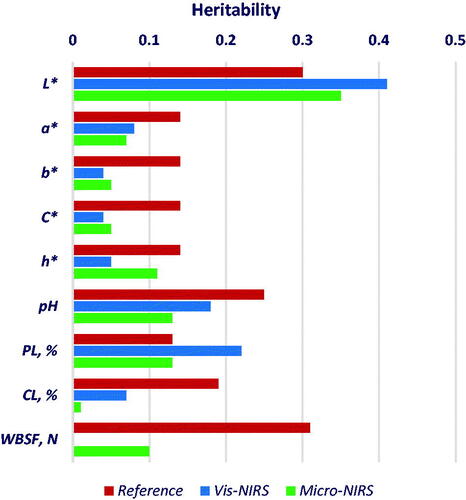
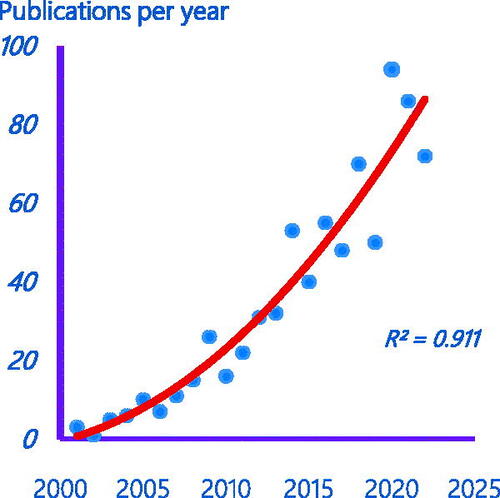
Figures & data
Table 1. Importance of genetic variability (Biodiversity) and genetic improvement (Genetics) for the phenotypic variability of the major traits affecting the product, process and overall quality of beef meat.
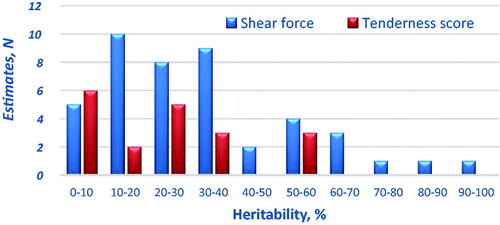
Table 2. Summary of the descriptive statistics and heritability estimates of beef quality traits obtained from studies carried out in the last 20 years (22 studiesTable Footnotea, 31 populations and 161,574 head).
Table 3. Summary of the major differences in F2 crossbred steers and heifers obtained from F1 bulls and dams (heterozygotes mh/+) sired by Piemontese bulls (homozygotes mh/mh) according to the number of mutated mh alleles inherited from the Piemontese grandsires.