Figures & data
Figure 1. Primary structure diagrams of Sis1 and Hdj1 (DNAJB1) expression constructs described in this manuscript. Protein regions are denoted using the following notation: J, J-domain; G/F, glycine/phenylalanine-rich region; G/M, glycine/methionine-rich region; CTD1/2, C-terminal peptide-binding domains I and II; DD, dimerization domain.Citation1,27 Dashed lines indicate where a region had been deleted.
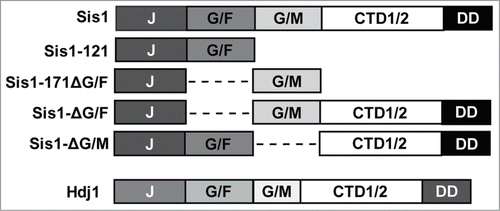
Figure 2. Hdj1 is deficient in replacing Sis1 in the curing of [PSI+] by Hsp104 overexpression. (A & B) [PSI+] cells of the W303 (panel A) or 74D-694 (panel B) genetic backgrounds bearing strong [PSI+] variants ([PSI+]STR, [PSI+]Sc4, or [PSI+]VH) and expressing Hdj1 in place of Sis1 (left columns) were transformed by a plasmid overexpressing Hsp104 ([pRS426-GPD-HSP104]) that normally cures all variants of [PSI+] (right columns). Color phenotype assays are shown for representative transformants (n ≥ 10 for each variant) following passage on media selecting for the Hsp104 overexpression plasmid. (C) Maintenance or loss of [PSI+] in cells shown in panel B was also confirmed by semi-denaturing detergent agarose gel electrophoresis (SDDAGE). Detergent resistant Sup35 aggregates indicative of the presence of [PSI+] were resolved by SDDAGE and visualized by immunoblot analysis using an antibody specific for Sup35. Control [psi−] cells were included for comparison. (D & E) Protein expression levels in cells from the 74D-694 genetic background were measured for Sis1 (panel D) and Hsp104 (panel E). Cell extracts from isolates in panel B were subjected to immunoblot analysis using antibody specific for Sis1 or Hsp104. A band cross-reacting with each antibody is shown as a loading control. Control [psi−] cells were included for comparison. Sis1 and Sup35 antibodies and methods for SDDAGE, SDSPAGE, and immunoblotting are the same as described in Harris et al.Citation1
![Figure 2. Hdj1 is deficient in replacing Sis1 in the curing of [PSI+] by Hsp104 overexpression. (A & B) [PSI+] cells of the W303 (panel A) or 74D-694 (panel B) genetic backgrounds bearing strong [PSI+] variants ([PSI+]STR, [PSI+]Sc4, or [PSI+]VH) and expressing Hdj1 in place of Sis1 (left columns) were transformed by a plasmid overexpressing Hsp104 ([pRS426-GPD-HSP104]) that normally cures all variants of [PSI+] (right columns). Color phenotype assays are shown for representative transformants (n ≥ 10 for each variant) following passage on media selecting for the Hsp104 overexpression plasmid. (C) Maintenance or loss of [PSI+] in cells shown in panel B was also confirmed by semi-denaturing detergent agarose gel electrophoresis (SDDAGE). Detergent resistant Sup35 aggregates indicative of the presence of [PSI+] were resolved by SDDAGE and visualized by immunoblot analysis using an antibody specific for Sup35. Control [psi−] cells were included for comparison. (D & E) Protein expression levels in cells from the 74D-694 genetic background were measured for Sis1 (panel D) and Hsp104 (panel E). Cell extracts from isolates in panel B were subjected to immunoblot analysis using antibody specific for Sis1 or Hsp104. A band cross-reacting with each antibody is shown as a loading control. Control [psi−] cells were included for comparison. Sis1 and Sup35 antibodies and methods for SDDAGE, SDSPAGE, and immunoblotting are the same as described in Harris et al.Citation1](/cms/asset/99b68984-3cbd-4bc9-8845-0c8cefdb4f73/kprn_a_1020268_f0002_c.gif)
