Figures & data
Figure 1. Schematic representation of PrPC structural determinants. Highlighted: N-terminal domain (red line), globular domain (blue line), octarepeat region (OR) with octapeptide sequences (see main text), α-helices (H1, H2 and H3), β-strands (S1, S2), N-linked glycosylation sites (CHO), disulfide bridges (S-S) and glycosylphosphatidylinisotol (GPI) anchor.
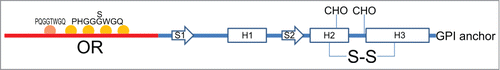
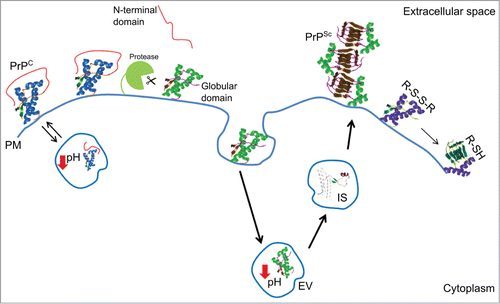
Figure 2. Cartoon of the major structural determinants and environmental key factors in prion protein folding and stability. Abbreviations correspond to: cellular prion protein (PrPC); prion (PrPSc); highly enthalpically stable intermediate state (IS); plasma membrane (PM); endosomial vesicles (EV); disulfide bridge (R-S-S-R); free thiol (R-SH).