Figures & data
FIGURE 1. Recombinant prion protein from with human, bovine and porcine sequence was fibrillated in vitro under near native conditions as described in Nyström & Hammarström (2015).Citation12 The bars represent lag times in minutes for unseeded (black columns) and seeded with 1% preformed fibrils of all the included sequences (blue for human, green for bovine and pink for porcine).
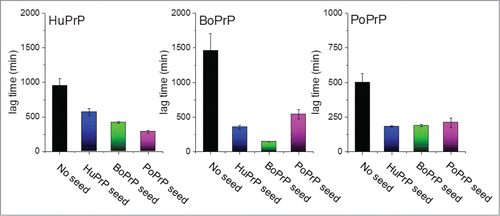
FIGURE 2. Structure and sequence comparisons. (a) Overlaid NMR structures of human, bovine and porcine PrP (blue: HuPrP PDB code 1QM2; green: BoPrP PDB code 1DWY; pink: PoPrP PDB code 1XYQ). (b) Sequence alignment with residues unique for one of the 3 species indicated with color code as in a)

FIGURE 3. Schematic model of prion strain adaptation. (Model adapted from Collinge and Clarke 2007 and Sandberg et al 2011, 2013.Citation31,49,50) The red horizontal line indicates the tolerance threshold for prion toxicity for the respective model, the green vertical line indicates normal lifespan/experimental termination for the mice. The black curves indicate increase in prion titer over time upon prion inoculation. (a) BSE and classical scrapie in wild type mice according to Bruce et al.Citation23 (b) BSE, classical scrapie and Nor98 scarpie in PoTg001 mice according to Espinosa, Torres et al. (2009, 2014).Citation25,26
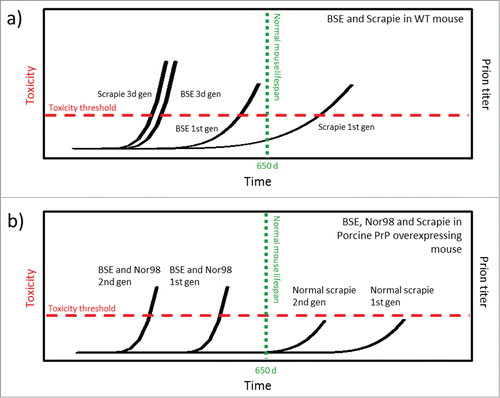
FIGURE 4. Potential prion strain adaptation in pig. The red horizontal gradient indicates the hietherto unkown prion toxicity tolerance threshold for pigs, the blue vertical line indicates normal slaughter age for industrial pig farming, the green vertical line indicates the normal lifespan of a breeding sow in industrial scale pig farming, orange areas indicate window of neurotoxic prions before onset of clinical disease (dark orange indicates subclinical BSE as reported by Wells et al,Citation16 pale orange indicate hypothetic outcome of PSE and strain adaptation. On the outmost right a potential subclinical sporadic PSE.
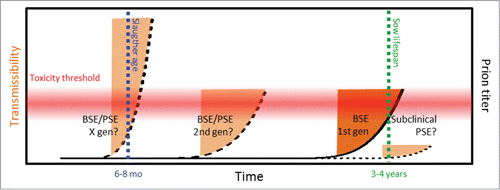
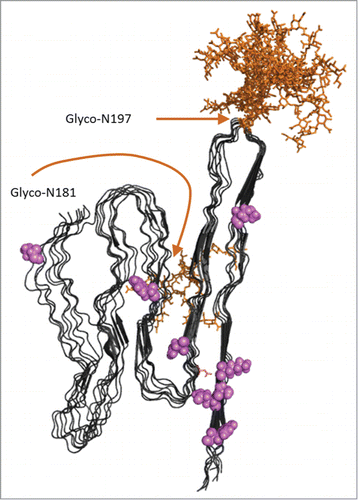
FIGURE 5. Model of APrP fibrillar structure displayed as 8 in register parallel beta-sheet conformations as suggested by Groveman et al 2014 based on hamster PrP 90–231.Citation41 Glycosylations (complex glycans) were added in silico using GlyProt.Citation54 Both N197 and N181 were distinguished as putative glycosylation sites by GlyProt. However, N181 was only accessible for glycosylations for one PrP chain at the end of the “elongating fibril” as shown in the back side of the model. N197 was accessible for glycosylation in all 8 PrP chains. Glycans are indicated in orange. Amino acid positions unique for PoPrP relative to HuPrP and BoPrP () are indicated in pink.