Figures & data
FIGURE 1. (See previous page).Analysis of immortalized bovine fibroblasts. (A) Bright-field pictures of in vitro cultured bovine fibroblasts. BFs at passage 7 were used as a control (a), and BBFs (b) and BTBFs (c) represented non-morphological changes at passage 42 compared with the early passage of BFs. (B) The population doubling time of immortalized bovine fibroblasts. Senescent cells did not show any cellular growth after 80 days of primary culture, but immortalized cells were populated continuously after the BFs' senescence. (C) The doubling time was only increased in BBFs as their passage increased, and they arrived at cellular senescence after passage 43 (b). (D) Clonogenic assay of each cell line; single colonies of BFs, BBFs, and BTBFs were stained using crystal blue, and the number of colonies in each group were counted (E). (F) Karyogram of BTBFs at passage 52. BTBFs have normal karyotype (b) like BFs (a). (BFs: Primary cultured bovine fibroblasts, BBFs: Bmi-1-introduced bovine fibroblasts, BTBFs, Bmi-1+hTert-introduced bovine fibroblasts).
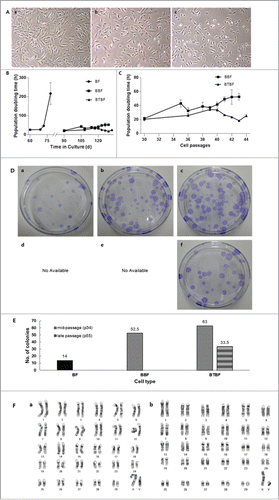
FIGURE 2. Embryonic development, telomerase activity, and gene expression. (A) Development rate of cloned embryos of BFs, BBFs, and BTBFs. Reconstructed embryos' fusion, cleavage, and blastocyst formation rates were estimated one hour after electric fusion, on day 2, and on day 7, respectively. (B) Among the blastocysts in each group, bastocyst formation ratio was differently observed on Day 7 and Day 8. (C) The cell number of the blastocysts in each group was counted after differential staining, and the ICM/TE ratio was calculated. (D) Telomerase activities were measured in control cell lines (passage 5 (BF5), passage 20 (BF20), BBF lines (passage 20 (BBF20), passage 40 (BBF40)), BBBF lines (passage 20 (BTBF20), passage 40 (BTBF 60)). In the BTBF60, telomerase activity was detected in the highest level. (E) p53 gene expression by quantitative RT-PCR was measured in various cell passages of BFs, BBFs, and BTBFs. (BFs: Primary cultured bovine fibroblasts, BBFs: Bmi-1-introduced bovine fibroblasts, BTBFs, Bmi-1+hTert-introduced bovine fibroblasts).
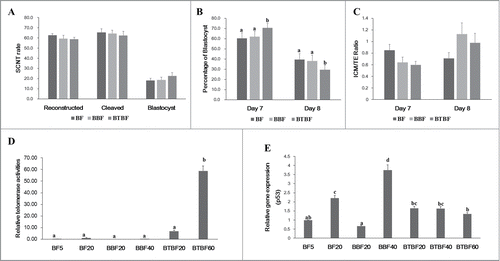
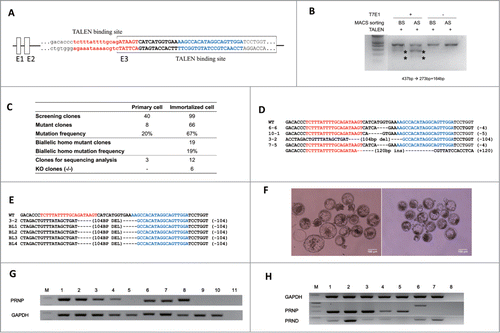
FIGURE 3. Knockout of the PRNP gene in immortalized bovine fibroblast cell lines. (A) TALEN target site on exon 3 of bovine PRNP gene. Recognition sequences for left TALEN (red) and right TALEN (blue) are flanking a 12-bp spacer (bold). (B) Enrichment of PRNP-modified cells via a surrogate reporter-mediated MACS assessed by T7E1 assay. The cleavage products by T7E1 due to the TALEN-induced mutation on the PRNP gene are depicted with asterisks. (C) Summary of the PRNP knockout results in primary and immortalized bovine fibroblasts. (D) The genotype of the PRNP mutation in 4 representative knockout clones. Note that identical mutations were introduced to both alleles in selected clones, except for 7–5. (E) Mutant allele analysis of cloned blastocysts derived from a knockout immortalized cell line. (F) Representative pictures of cloned blastocysts from immortalized cells (left) and knockout cells (right). (G) Expression of PRNP mRNA in knockout blastocysts (M: DNA ladder, 1: Spinal cord, 2: Cumulus cells, 3: Primary cells, 4: Immortal cells, 5: KO immortal cells, 6: in vitro fertilized blastocysts, 7 and 8: Blastocysts from immortalized cells, 9 and 10: Blastocysts from knockout immortalized cells, 11: Negative controls). (H) PRNP and PRND expression by RT-PCR in PRNP KO cell lines. (M: DNA ladder, 1: Spinal cord, 2: Testis, 3: Primary cells, 4: Immortal cells, 5: KO cells 6–6, 6: KO cells 7–5, 7: KO cells 3–2, 8: Negative controls).