Figures & data
TABLE 1. Phenotypic characteristics of goat breeds in Pakistan under study.
TABLE 2. Polymorphism observed in PrP gene in Pakistani goat breeds of meat and milk potential.
TABLE 3. Level of polymorphism summarized according to percentage observed at each nucleotide position.
TABLE 4. Single nucleotide polymorphisms identified in the PrP region of animals all over the world.
TABLE 5. Comparison between studied PrP sequences of animals all over the world with indeginous genotypes of goat breeds of Pakistan. Entries in bold and underlined format in the reported column were also determined as amino acid polymorphisms in studied breeds. Similarly, one entry placed the 3rd spot in the right most column was the novel polymorphism detected in indigenous goat population.
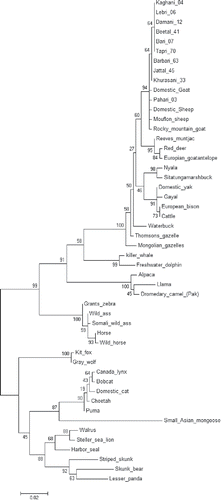
FIGURE 2. NJ Phylogenetic tree constructed using maximum likelihood method between 11 Pakistani goat breeds and 40 other mammalian species. Bootstrap value was kept 1000.
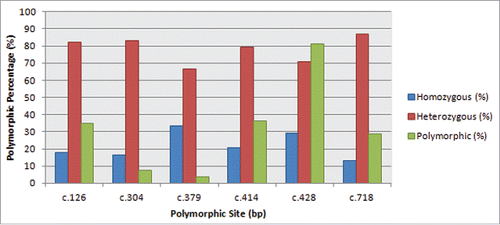
FIGURE 3. Barry and Hartigan's relative rate test for the comparison of evolutionary distance between PrP homologs in different breeds/species.
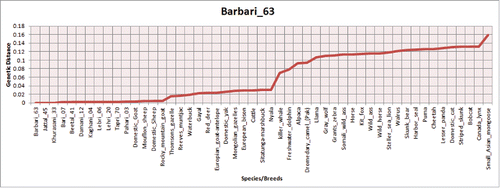
FIGURE 4. Multidimensional Scaling plot of genetic distance between populations (Basques – closed circles, Iberia – open circles, Europe – closed squares, Middle East – cross, Caucasus – closed triangles, North Africa – open triangles). The first 2 axes account for highest genetic variation present in the sample and can be demonstrated that the plot is an accurate representation of the genetic distance matrix. The small ruminant group cluster together on the bottom-left side of the plot, near their bigger ruminant neighbors represented on the upper left place. The canine populations are found near the bottom-right of plot, differentiated from the other 2 ruminant groups.
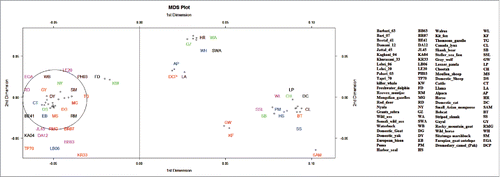
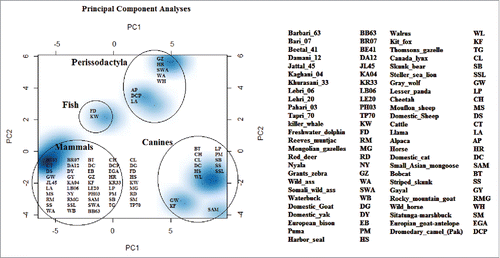
FIGURE 5. Principal component analyses resolve the genetic distance matrix of selected species into PC1, PC2, and PC3 and so on. Out of these, PC1 and PC2 represents large proportion of variation in the dataset and are plotted. PCA results enhanced consistency in our data set as all 3 groups make their spots according to their phylogenetic links. Here PC1 is quoting largest variation in the dataset as compared to PC2. So neighborhood has been revealed between cattle and goat. In that study central nervous system specific human enhancers exhibit 3 distinctive and internally compact clusters exposing the interactive pattern of TFBSs. The control data set of human non-conserved and non-coding elements do not present any vivid cluster structure for the 14 TFBSs that formed internally closely packed clusters in CNS specific enhancers, thus elucidating the significance of clusters in panel A. TFBSs are color coded.