Figures & data
FIGURE 1. Localization of murine-caprine chimeric PrPC molecules harboring either the wild type mature caprine PrP sequence or each one of the tested caprine alleles, within 22LN2a#58 cells. Chimeric murine –caprine PrP molecules consisting of the mature caprine PrP sequence and harboring either the wild type (wt), or the Asp-146, His-154, Gln-211 and Lys-222 variants were expressed in scrapie-infected 22LN2a#58 cells and detected by indirect immunofluoresence (green). Primary antibody: P4 (dilution 1/250), secondary antibody: GAM-FITC (dilution 1/1,000). Nuclei were counter-stained with propidium iodide (red). Primary antibody specificity was determined by processing untransfected cells (untr) as described before. Secondary antibody specificity was determined by processing transfected cells expressing the wild type chimeric PrP only with the secondary antibody (neg). Samples were observed under a Confocal Laser Scanning Microscope (Zeiss, Observer Z1), using the following filters: emission 488 nm - detection 515/530 nm (green), emission 543 nm - detection 570 nm (red). Magnification: x40. Scale bar: 20 μm. Peri-nuclear (white arrows), plasma membrane (white dashed arrows), neurite and neuronal axon localization (star) was detected.
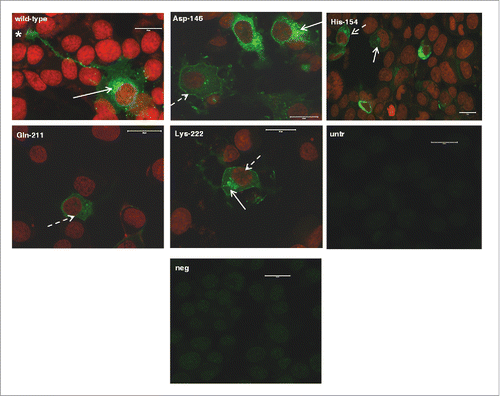
FIGURE 2. Murine-caprine chimeric PrP molecules harboring the wild type mature caprine PrP sequence are converted to partially resistant to Proteinase K treatment moieties in the murine scrapie-infected 22LN2a#58 cells. Western blot analysis on cell lysates from scrapie-infected (22LN2a#58) or non-scrapie infected (N2a#58) cells expressing murine-caprine chimeric PrP molecules harboring the wild type mature caprine PrP sequence after transient transfection with the corresponding vector. Two days after transfection cells were lysed; lysates were either treated with Proteinase K (PK+) or not (PK-), and proteins were methanol precipitated. Protein pellets were resuspended in sample buffer, analyzed by SDS-PAGE, electrotransferred on a PVDF membrane and probed with the sheep/goat PrP-specific L42 antibody (dilution 1/10). The secondary antibody (GAM-HRP) was used at a 1/10,000 dilution. The blot was developed on autoradiography film, with enhanced chemiluminesence (ECL). For PK treatment 0.75 μgs PK/mg total protein were used for 1 h at 37°C, with agitation; reaction was stopped with addition of PMSF. Analyzed protein loads correspond to 10 μgs for PK(-) samples and to the equivalent of 100 μgs before PK-treatment for PK(+) samples. Pertinent Molecular Weight Markers are indicated. An L42 immunopositive band at ∼25 kDa is detected in PK-treated lysates from scrapie-infected cells, while it is absent from scrapie-free cells, indicating that the exogenously expressed PrP undergoes conformational rearrangement in the scrapie-infected cells, which results in partial resistance to PK.
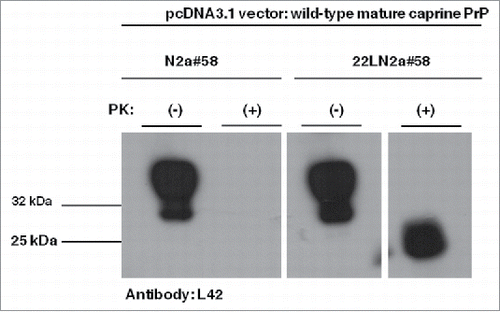
FIGURE 3. Goat specific Proteinase K-resistant moieties produced in the 22LN2a#58 cells are different from PrPSC molecules of naturally occurring goat scrapie cases. Western blot analysis on cell lysates from scrapie-infected (22LN2a#58) or non-scrapie infected (N2a#58) cells expressing murine-caprine chimeric PrP molecules harboring the wild type mature caprine PrP sequence. The same samples as in were subjected to SDS-PAGE, electrotransferred onto a PVDF membrane and probed with the P4 antibody (1/250 dilution), which recognizes a PrP region close to the PK cleavage site. Pertinent Molecular Weight Markers are indicated on the left. Absence of a P4 immunopositive band at ∼25 kDa in PK-treated lysates from both scrapie-infected and scrapie-free cells, indicates that the goat specific Proteinase K-resistant moieties produced in the scrapie-infected cells () are different from the PrPSC molecules detected in natural scrapie cases.Citation29
FIGURE 4 (see next page). Murine-caprine chimeric PrP molecules harboring the Asp-146, His-154 and Gln-211 caprine PrP variants display reduced conversion efficiency to partially Proteinase K- resistant moieties in murine scrapie-infected 22LN2a#58 cells. (A) Representative Western Blot on cell lysates from 22LN2a#58 cells, expressing murine-caprine PrP chimeras carrying either the wild type mature caprine PrP sequence or the Asp-146, His-154, Gln-211 and Lys-222 variants, after transient transfections with the corresponding vectors. Cells were lysed 2 d after transfection and lysates were either subjected to PK-treatment (PK+) or not (PK-). Protein samples were analyzed by SDS-PAGE, electrotransferred onto a PVDF membrane and probed with the sheep/goat PrP-specific L42 antibody (1/10 dilution). The secondary antibody (GAM-HRP) was used at a 1/10,000 dilution. For development, enhanced chemiluminescence (ECL) was used. Protein loads correspond to lysate volumes containing similar amounts of the exogenously expressed proteins. PK treatment was preformed with 0.75 μgs PK/mg total protein, at 37°C, for 1 h, with agitation, and was stopped with the addition of PMSF. Protein loads for PK(+) samples are 10-fold higher than the corresponding loads of PK(-) samples. PK treated samples were subjected to methanol precipitation before analysis. Pertinent Molecular Weight Markers are indicated. Immunopositive L42 bands of ∼25 kDa in PK-treated samples correspond to partially PK-resistant moieties. (B) Histograms depicting the mean and standard errors of the tested alleles' conversion efficiencies relative to wild type, determined by 3 independent experiments. Conversion efficiencies were estimated as the ratio of L42 immunopositive band intensities at ∼25 kDa in PK-treated samples (PrPSC) to immunopositive band intensities at ∼32 kDa in non-PK treated samples (total PrP, PrPC and PrPSC), and were expressed as a percent of the conversion efficiency of the most susceptible allele (wild type). Band intensities were determined by densitometric analysis performed on corresponding Western blots (A), using the ImageJ software. Unpaired student t-test was used for statistical analysis. **: p <0.01, ns: not statistically significant
FIGURE 5. Hydrogen bond formation prediction within caprine PrP variants harboring the tested alleles compared to wild type. (A) Structural model of the caprine PrP globular domain, based on ovine PrP crystallographic data [PDB entry 1uw3]Citation30 and visualized using the pdb.viewer software. Amino acid residues corresponding to allelic variants of interest are shown in red. Helixes are shown in orange and β-sheet in green. (B-E) Snapshots of the caprine PrP molecule, depicting hydrogen bonds formed in wild type and in variants harboring amino acid substitutions. Residues of interest are shown in red; hydrogen bonds are depicted as dashed light green lines. (B) Prediction of the Asp-146 allele effect on hydrogen bond formation relative to wild type (wt). No changes are predicted by the substitution of Asparagine by Aspartic acid at position 146, (C) Prediction of the His-154 allele effect on hydrogen bond formation compared to wild type (wt). No changes in hydrogen bonds formation are predicted by the substitution of Arginine by Histidine at position 154, (D) Prediction of the Gln-211 allele effect on hydrogen bond formation compared to wild type (wt). Substitution of Arginine by Glutamine at residue 211, diminishes the hydrogen bond formed between Arg-211 and His-143 and ablates the connection between helix3 and the S1H1 loop. Note that for easier depiction, parts of helix3 (orange surfaces) are not displayed, (E) Prediction of the Lys-222 allele effect on hydrogen bond formation relative to wild type (wt). Substitution of Gln-222 by Lysine, abolishes the hydrogen bond formed between Gln-222 and Gln-175. For easier depiction, parts of helix3 are not displayed.
![FIGURE 5. Hydrogen bond formation prediction within caprine PrP variants harboring the tested alleles compared to wild type. (A) Structural model of the caprine PrP globular domain, based on ovine PrP crystallographic data [PDB entry 1uw3]Citation30 and visualized using the pdb.viewer software. Amino acid residues corresponding to allelic variants of interest are shown in red. Helixes are shown in orange and β-sheet in green. (B-E) Snapshots of the caprine PrP molecule, depicting hydrogen bonds formed in wild type and in variants harboring amino acid substitutions. Residues of interest are shown in red; hydrogen bonds are depicted as dashed light green lines. (B) Prediction of the Asp-146 allele effect on hydrogen bond formation relative to wild type (wt). No changes are predicted by the substitution of Asparagine by Aspartic acid at position 146, (C) Prediction of the His-154 allele effect on hydrogen bond formation compared to wild type (wt). No changes in hydrogen bonds formation are predicted by the substitution of Arginine by Histidine at position 154, (D) Prediction of the Gln-211 allele effect on hydrogen bond formation compared to wild type (wt). Substitution of Arginine by Glutamine at residue 211, diminishes the hydrogen bond formed between Arg-211 and His-143 and ablates the connection between helix3 and the S1H1 loop. Note that for easier depiction, parts of helix3 (orange surfaces) are not displayed, (E) Prediction of the Lys-222 allele effect on hydrogen bond formation relative to wild type (wt). Substitution of Gln-222 by Lysine, abolishes the hydrogen bond formed between Gln-222 and Gln-175. For easier depiction, parts of helix3 are not displayed.](/cms/asset/e3cf6e52-8445-4670-ba6b-1ad846c5801d/kprn_a_1199312_f0005_oc.gif)
