Figures & data
FIGURE 1. Structural and functional organization of the Saccharomyces cerevisiae Sup35 protein. (A) Domain organization of Sup35. Designations N, M and C refer to the Sup35N (N-proximal), Sup35M (middle) and Sup35C (C-proximal) regions, respectively. Numbers correspond to amino acid positions. (B) Hypothetical model of tertiary structure of the non-prion isoform of Sup35. N domain is intrinsically unfolded, structure of M domain is unknown (shown as unfolded), the C domain structure is based on cryo-electron microscopy analysis (EMDB accession ID: 4crn).Citation89
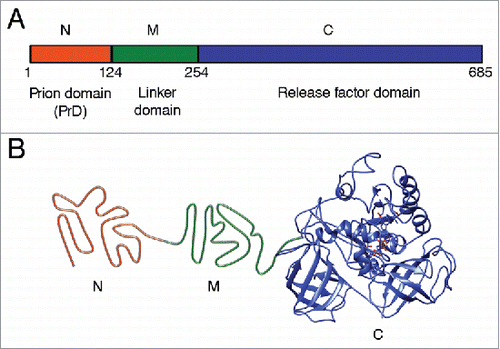
TABLE 1. Identity of amino acid sequences for different domains of Sup35 protein from Saccharomyces paradoxus and S. uvarum in comparison to S. cerevisiae.
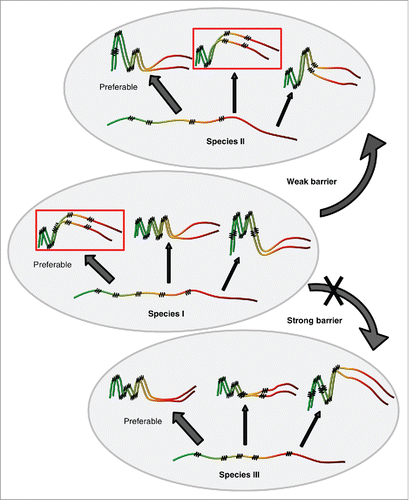
FIGURE 2. Transmission of prion state to a divergent protein. Large ellipse on the left represents yeast cell. Gray area represents the cellular compartment or quality control protein deposit containing aggregating proteins. Dotted ellipse indicates the specific interacting protein molecules that are considered in more detail and at larger magnification in the images to the right. Nonprion form of heterologous Sup35 prion domain protein is represented as unstructured black tangle while prion polymers are shown by light (orange) (pre-existing “donor” prion) or dark (the newly formed heterologous prion) pleated lines demonstrating in-register parallel β-sheet architecture. Short crosscut lines indicate positions of interactions between β-sheets. Prion domains with high identity of amino acid sequences in the regions, corresponding to the cross-β core of a donor protein, can convert each other to prion form with high frequency, while prion proteins with lower identity typically cannot adopt the donor strain conformation and form nonprion aggregates; however, in rare cases, the recipient protein is converted to a prion conformation, that is only partly collinear to the original donor conformation, and partly different from it.
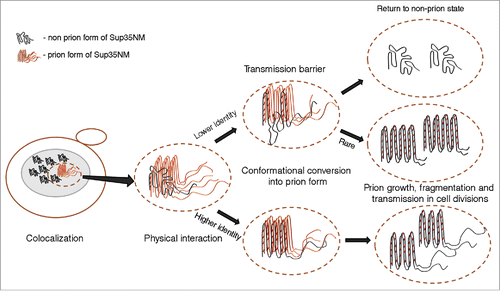
FIGURE 3. Model for cross-species prion transmission. Large gray ellipses correspond 3 different yeast species. Non-prion form of PrD domain of Sup35 protein is shown as wavy line, while prion polymers are shown by pleated lines demonstrating in-register parallel β-sheet architecture. Zigzags correspond to turns between β-strands. Prion strains formed by one and the same protein differ from each other by both size/location of cross-β regions, and positions of turns. Due to differences in amino acid sequences, homologous proteins from different species can generate different spectra of strains. Each species has a preferable strain conformation (pointed to by a thick arrow), and different strains may have different preferable conformations. When prion proteins from different species can adopt similar conformations (as in examples within the rectangles), and a donor protein prefers such a conformation (or is present in such a conformation in the specific experiment), cross-species prion transmission may occurs relatively efficiently, and prion species barrier is weak (as shown for species I and II on this Figure). When prion proteins from different species do not produce strains of identical or similar conformations, strong species barrier is detected (as shown for species I and III on this Figure).