Figures & data
Figure 1. Dilution-response profiles of the kits using the brain homogenate of a C-BSE cow. Raw data were plotted as the mean ± SEM (standard error of the mean) from a set of triplicate wells. The dotted lines indicate thresholds of positivity defined by the manufacturers' protocols. Non-linear curve fitting was applied to the row data using a four-parameter logistic model. (A) TeSeE®, (B) FRELISA®, (C) NippiBL®, and (D) BetaPrion®.
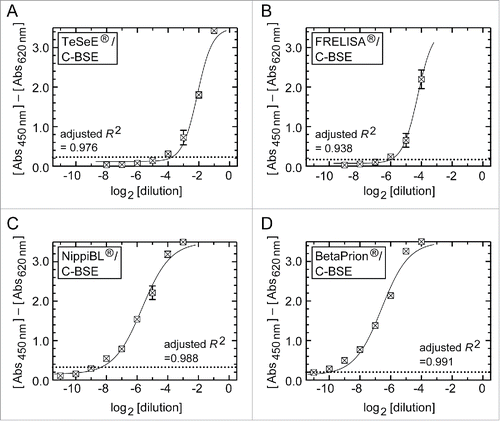
TABLE 1. Detection limits of the ELISA tests for C- and L-BSE samples.
Figure 2. Dilution-response profiles of the kits using the brain homogenates of L- and H-BSE cows. Raw data were plotted as the mean ± SEM from a set of triplicate wells. The dotted lines indicate thresholds of positivity defined by the manufacturers' protocols. Non-linear curve fitting was applied to the row data using a four-parameter logistic model. (A – D) TeSeE®, FRELISA®, NippiBL®, and BetaPrion® tested on the samples prepared from the L-BSE cow, respectively. (E) NippiBL® tested on the samples prepared from the H-BSE cow.
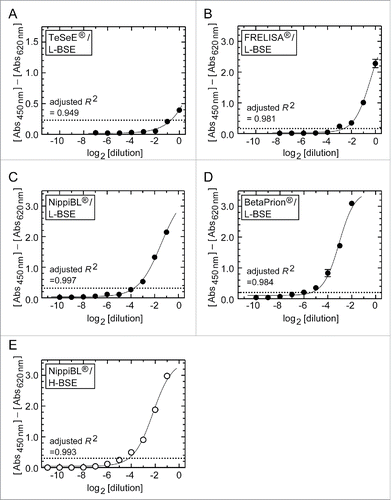
Figure 3. Western blot analysis after PK digestion to determine the relative amounts of PrPSc in the stock homogenates of the brain. (A) The stock homogenates of the brains of the C-, L-, and H-BSE cows were digested by PK, and aliquots of volumes of the digests corresponding to the indicated weights of tissues were subjected to Western blot analysis. PrPSc was detected using anti-PrP antibody 12F10, with the aid of a chemluminescent detection reagent and a cooled CCD camera imaging system. The letters non-, mono-, and di- denote the non-, N-mono-, and N-di-glycosylated forms of PrPSc. (B) Signal intensities of the non-, mono-, and di-glycosylated forms of PrPSc in each lane in (A) were measured by ImageGuage software, combined as a total signal intensity of PrPSc, and plotted in relative magnitude by taking that of 50 μg of the C-BSE brain tissue as 10.0.
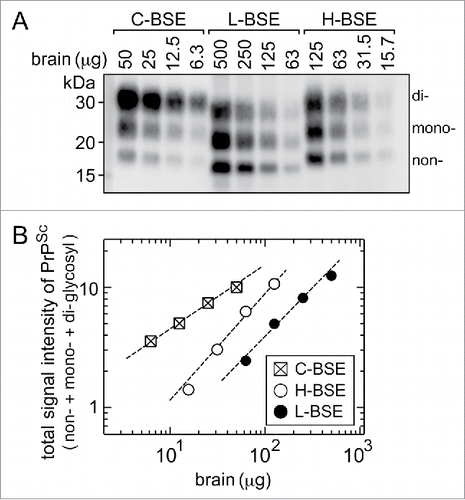
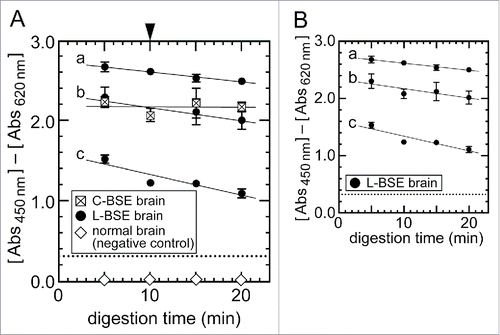
Figure 4. Effect of the sample digestion condition of NippiBL® on detection of the L-BSE prion. (A) Brain samples of the C-BSE and L-BSE cows were digested according to the manufacturer's protocol but for an extended time. For the L-BSE brain, samples of three different dilutions by normal brains were tested (a: 2−0.6 dilution, b: 2−1 dilution, c: 2−2 dilution). The arrowhead at the top of (A) indicates the digestion time set by the protocol (10 min). Data were plotted as the mean ± SEM from duplicate wells. The dotted lines indicate the thresholds of positivity defined by the protocol. The samples of normal brain (i.e., negative control) gave rise to negative signals throughout the assay. (B) Only data on the brain samples of the L-BSE cow in (A) were plotted for clarity.