Figures & data
Figure 1. Association with Las17 is required for prion induction by Lsb2. A. Lsb2 cannot induce formation of prion [PSI+] by overexpression of Sup35 in the strain depleted of Las17 (las17Δ). Depletion of Las17 does not affect formation of [PSI+] by overexpression of Sup35 in the presence of prion aggregates of Rnq1 protein ([RNQ+]). [PSI+] formation is manifested by growth on SD-Ade medium. B. Total levels of Lsb2 protein are slightly decreased in Las17 strain as detected by western blotting. Different time of Lsb2 induction from plasmid copper inducible promoter is indicated. C. Wild type Lsb2 and more stable ubiquitination deficient mutants (K80; K80R, K41R; P124A, P125A) do not form detergent resistant amyloid-like aggregates in the strain depleted of Las17 as detected by SDD-AGE. Lsb2 W91S mutant unable to bind Las17 does not form amyloid-like aggregates in the presence of Las17 (WT).
![Figure 1. Association with Las17 is required for prion induction by Lsb2. A. Lsb2 cannot induce formation of prion [PSI+] by overexpression of Sup35 in the strain depleted of Las17 (las17Δ). Depletion of Las17 does not affect formation of [PSI+] by overexpression of Sup35 in the presence of prion aggregates of Rnq1 protein ([RNQ+]). [PSI+] formation is manifested by growth on SD-Ade medium. B. Total levels of Lsb2 protein are slightly decreased in Las17 strain as detected by western blotting. Different time of Lsb2 induction from plasmid copper inducible promoter is indicated. C. Wild type Lsb2 and more stable ubiquitination deficient mutants (K80; K80R, K41R; P124A, P125A) do not form detergent resistant amyloid-like aggregates in the strain depleted of Las17 as detected by SDD-AGE. Lsb2 W91S mutant unable to bind Las17 does not form amyloid-like aggregates in the presence of Las17 (WT).](/cms/asset/feec79b9-d414-4559-8b28-83b042bf2cda/kprn_a_1328342_f0001_oc.gif)
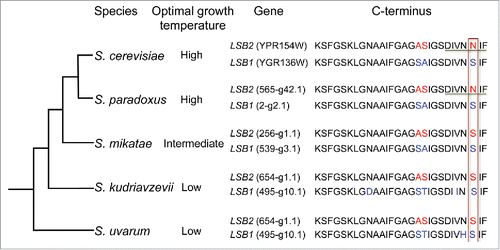
Figure 2. Prion–inducing activity of Lsb2 coincides with yeast adaptation to higher growth temperature. Schematic shows phylogenetic relationships among some members of the Saccharomyces sensu stricto genus. The preferred growth temperature of each species is indicated.Citation74,75 CLUSTALW-formatted multiple sequence alignment of C-terminal of Lsb1/Lsb2 is shown. Difference in amino acids is indicated in red (Lsb2) and blue (Lsb1). Residue essential for prion induction is bordered. Amyloid stretch hexapeptide is underlined.

![Figure 3. Model “Metastable prion [LSB+] controls stress memory.” Subpopulation of cells with [LSB+] maintains a memory of stress and is better adapted to it. See comments in the text.](/cms/asset/8d25ee94-daad-4416-bc35-7204511fb47c/kprn_a_1328342_f0003_oc.gif)