Figures & data
Figure 1. Intracellular localization of PrP and neurotoxic properties in mammalian cells. Schematic presentation of PrP and mutants thereof. Dark rectangle: signal peptide; α-helical structure is indicated by helices, intrinsically disorder by a straight line; polygons represent N-linked glycosylation acceptor site; white rectangle: GPI anchor signal sequence. Please note that GPI signal peptides are predicted to adopt an alpha-helical structure. Neurotoxic PrP mutants linked to inherited prion diseases in humans (PrP-Q160X and PrP-Y145X) are characterized by deletions in the C-terminal globular domain and impaired import into the ER. Expression of cytoPrP in transgenic mice induces severe ataxia due to rapid cerebellar granule neuron degeneration. *It was described that also wildtype PrP can remain in the cytosol and interfere with cellular viability. For details see main text.
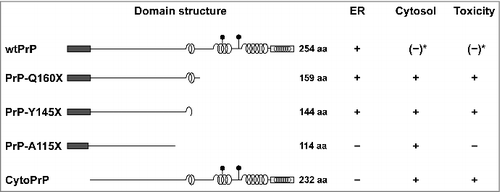
Figure 2. Conserved pathways for protein translocation in eukaryotes and prokaryotes. Schematic view of Sec61- and SecY-mediated secretion in eukaryotes and gram-negative bacteria. After targeting of the ribosome-nascent chain complex to the translocon the protein is translocated into the ER lumen or periplasm. In addition to the co-translational targeting pathway depicted in the scheme, substrates can also be targeted after they have been synthesized on free ribosomes in the cytosol (post-translational pathway).
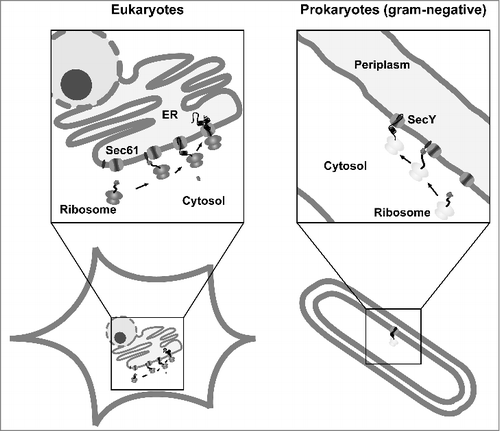
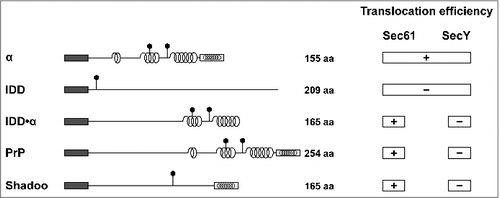
Figure 3. Selective impairment of the SecY complex to translocate secretory proteins with extended intrinsically disordered domains (IDDs) The scheme summarizes the translocation efficiency of different model substrates and natural occurring proteins, such as PrP and shadoo, into the ER of mammalian and yeast cells (Sec61) or the periplasm of E. coli (SecY). Dark rectangle: signal peptide; α-helical structure is indicated by helices, intrinsically disorder by a straight line; polygons represent N-linked glycosylation acceptor site; white rectangle: GPI anchor signal sequence. Please note that GPI signal peptides are predicted to adopt an alpha-helical structure.